Will Chen
@wchenomics
@DamonRunyon Fellow at Baker lab @UWproteindesign. PhD with @JShendure. mainly at https://bsky.app/profile/chenomics.bsky.social
Super excited to share that ENGRAM is out today! ENGRAM cells are programmed to write their histories into the genome, recording the intensity, duration, and order of biological events simultaneously. nature.com/articles/s4158…
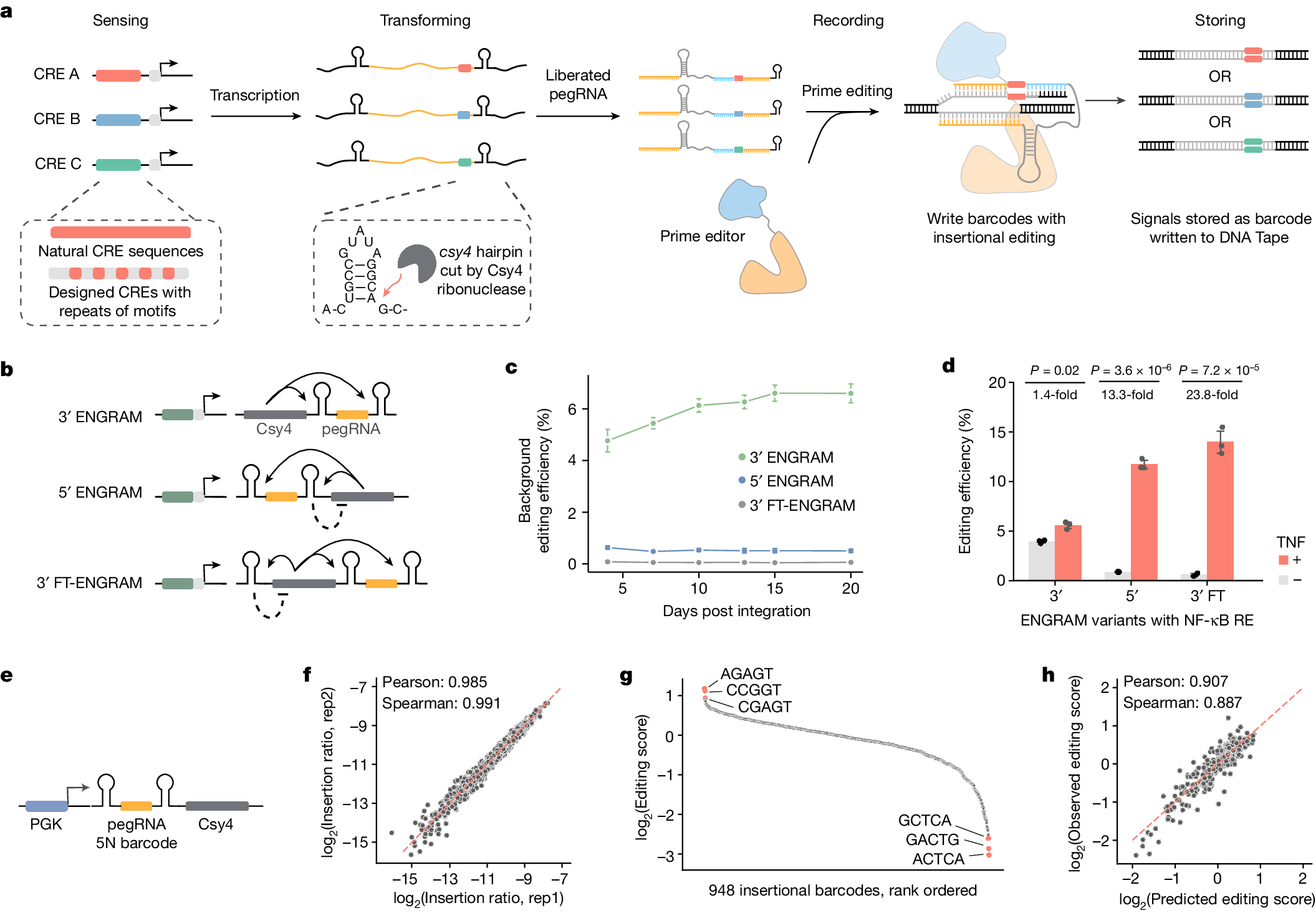
Congrats on this beautiful paper! @sudpinglay
Now out in @ScienceMagazine we present 'Genome-shuffle-seq': a method to shuffle mammalian genomes and characterize the impact of structural variants (SVs) with single-cell resolution in one experiment. science.org/doi/10.1126/sc…
Beautiful work! congrats @yodai_takei !
Excited to share our new paper out in @Nature revealing cell-type specific nuclear organization and its link to gene regulation using new spatial multi-omics technologies! nature.com/articles/s4158…
Congrats @JONA. such beautiful paper
I’m delighted to share our work on scrambling the human genome using prime editing, repetitive elements, and recombinases @ScienceMagazine! @ferreira_raph_ @LeopoldParts @ProfTomEllis, @geochurch @SangerInstitute science.org/doi/10.1126/sc… Lots of changes from the preprint. A🧵
Excited to finally share Perceptein, a PERCEPtron made of proTEINs: science.org/doi/10.1126/sc…
Finally out in @ScienceMagazine! Utilizing the cost-effective EasySci to profile 21 million single-cell transcriptomes across five life stages and diverse sexes/genotypes, we reveal aging as distinct, development-like transitions, with dramatic cell population changes in specific…
What if 1 million scRNA-seq libraries can be prepared within $1k? Here we go 👉 21 million cells from 623 mouse tissues, spanning 5 life stages & 3 genotypes, all in a SINGLE study by ONE remarkable student Zehao @Tommyz626 from our lab @RockefellerUniv! shorturl.at/epsz7
Bravo @adam_broerman! Really beautiful old-school design paper! this could open up new avenues for biosensor designs.
Excited to announce our new #proteindesign strategy for allosterically controlling the kinetics of protein-protein interactions! Read on for cool applications in cytokine signaling, biosensing, and protein circuits. biorxiv.org/content/10.110…
amazing work! congrats Mo!
Cute story alert!🧵@Caltech two clueless office mates (@piraner & I) created a tool to control cells with ultrasound. As postdocs (@UCSF & @UWproteindesign), we teamed up again (with @majo__duran & @AdamChazin) to build a tool for engineered cell signaling nature.com/articles/s4158…
beautiful work! have been waiting for this for a while! congrats @JoshTycko
Development of compact transcriptional effectors using high-throughput measurements in diverse contexts go.nature.com/3NMu00N
Beautiful paper and well summarized tweet! congrats @Yi__Fu
Excited to share my PhD work in @Agnelsfeir’s lab describing a method to engineer mitochondrial DNA deletions in human cells and our exploration of how cellular metabolism and transcription respond to deletion heteroplasmy. biorxiv.org/content/10.110… 1/19
Simply thrilled! Well-deserved recognition to David! Congrats! It's an exciting time to do protein design. Let's play more to modulate biology, or even create life
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”
Here’s our human protein-protein interactome. We mined the SRA, devised a new distillation dataset for protein complexes, trained a new version of RF2 to screen millions of protein pairs, and identify > 18k binary interactions. biorxiv.org/content/10.110…
I'm sharing my story in a difficult time for Palestinians and Lebanon, where we found refuge after being exiled from Haifa. I want to honor those who suffered the most and never got to live out their dreams—my grandparents, Jido Ali and Taita Khadejeh. I hope they’re proud (1/3)
The first episode of @BakerLabPodcast is live. Meet Mohamad Abedi, who went from being a Palestinian refugee to a postdoctoral researcher. His journey proves that science can be a creative and transformative process. Apple: podcasts.apple.com/us/podcast/the… Spotify: open.spotify.com/show/32pWWpKzO…
Congrats @FloChardon @TroyMcDiarmid ! beautiful paper!
Genomic tech dev is my favorite area of science, and today our paper describing our multiplex CRISPRa screening method to identify cell type specific regulatory elements is published! rdcu.be/dUnoq
Thank you so much @ChenMaggieSY for writing up the report.
For @JShendure and @wchenomics at @uwgenome, it’s about appreciating a cell’s genetic journey over time, not just its final destination. bit.ly/4e2LBg2
Amazing work! Congrats @Nobu_Hamazaki and @VinetteY and all involved. I learned a lot from both Nobu and Wei on gastruloid!
Excited to share our human RA-gastruloid model in @NatureCellBio! Early retinoic acid supplementation, followed by later Matrigel addition, robustly induces human embryonic morphologies and diverse cell lineages, including somitic, renal, cardiac, and neural cell types in…
Excited to announce our new paper on designing allosterically regulated protein assemblies from scratch, now published in @Nature! This collaborative effort included key contributions from @FloPraetorius, Abbas Idris, @AnnikaPhilomin, @cweidle1 and others! nature.com/articles/s4158…