Arvind S Pillai
@ArvindSPillai1
Washington Research Foundation Postdoctoral fellow, Institute of protein design. Formerly Phd student @UChicago doing evolutionary biochemistry. I also do art.
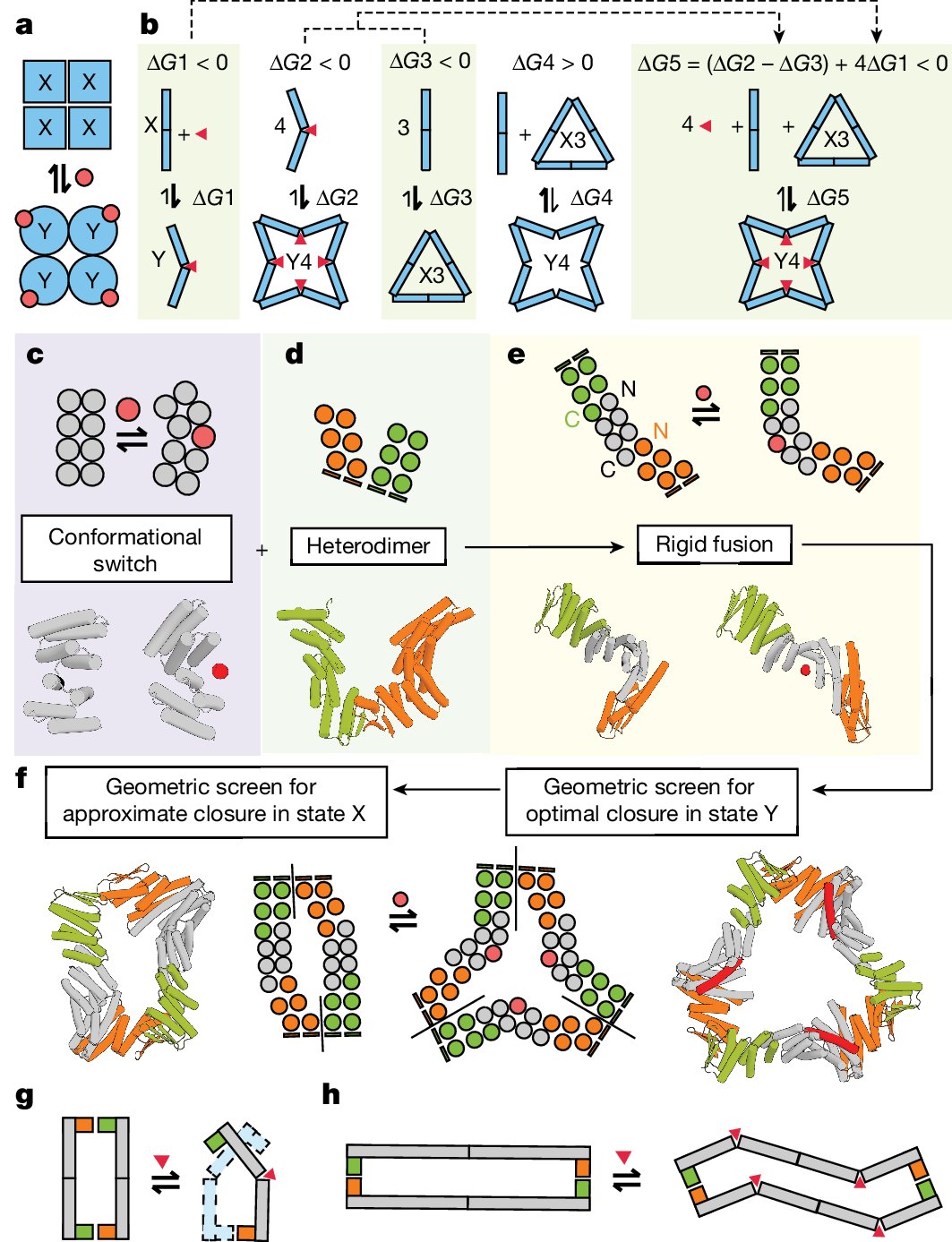
Excited to announce our new paper on designing allosterically regulated protein assemblies from scratch, now published in @Nature! This collaborative effort included key contributions from @FloPraetorius, Abbas Idris, @AnnikaPhilomin, @cweidle1 and others! nature.com/articles/s4158…
We’re proud to welcome two bold scientific minds to the Institute! Arvind Pillai, PhD, joins as Assistant Investigator exploring protein #evolution & design. Friederike Benning, PhD, joins as Stowers Fellow studying membrane complexity in #bacteria. More: bit.ly/3SCZQiM
Small proteins can be more complex than they look! We know proteins fluctuate between different conformations- but how much? How does it vary protein to protein? Can highly stable domains have low stability segments? @ajrferrari experimentally tested >5,000 domains to find out!
Happy National Draw a #Dinosaur Day! ✏️🦖 These masterpieces(?) were created during the 2023 National Fossil Day Livestream by Kallie Moore, Michelle Barboza-Ramirez, Blake de Pastino, and Gabriel Santos. Tag us in your dino drawings!
Read this awesome paper from @carway2nar about the biophysical and genetic mechanisms by which proteins evolve to assemble into highly specific assemblies of distinct molecular units. Proud to have contributed!
My first first-author paper is out! It was an amazing collaborative effort with a lot of amazing people including @ArvindSPillai1 and @JoeThorntonLab. Please read if you like things involving evolution and biochemistry. pnas.org/doi/10.1073/pn…
My first first-author paper is out! It was an amazing collaborative effort with a lot of amazing people including @ArvindSPillai1 and @JoeThorntonLab. Please read if you like things involving evolution and biochemistry. pnas.org/doi/10.1073/pn…
Excited and proud to have this paper published with @NatureComms today! Great project led by @DrJuanLMendoza at @UChicagoPME and @HHMINEWS. Excited for the future of the field - stay tuned for a full breakdown and walkthrough of our results! nature.com/articles/s4146…
Thrilled to see our work on protein nanocage design published in @Nature! These nanocages have exciting potential applications in drug delivery and vaccines. nature.com/articles/s4158… Thank you to Sangmin Lee for the opportunity to contribute to this amazing project!
New paper from us and @erblabs by @schulluc : embopress.org/doi/full/10.10…. It's about the evolution of essentiality. This question has fascinated me for a long time: How does a component that was previously unnecessary for some function become essential for it?
Read this amazing work from Adam & co on designing complexes whose kinetics of assembly and disassembly can be allosterically controlled with remarkable precision!
Excited to announce our new #proteindesign strategy for allosterically controlling the kinetics of protein-protein interactions! Read on for cool applications in cytokine signaling, biosensing, and protein circuits. biorxiv.org/content/10.110…
#CrispyShifty proteins mentioned - highlight of the announcement for me 😁 @ArvindSPillai1 @FloPraetorius
David didn’t just launch a field that's transforming science and soon, our lives—he did it while being an incredible mentor and a caring human. His dedication to his lab and students is just as worth celebrating. Cograts to David and to us at the @UWproteindesign!
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”
A reminder that proteins are highly dynamic molecules. ◼ We have made progress in measuring & predicting static protein structures, but the dynamics that animate life remain challenging to measure & model.
I think the actual "work" of academia is pretty fun and intrinsically rewarding. The discipline would be a lot better if we could all be more self-aware and intentional about reducing apathy, avoidance of hard but necessary things, and smug, self-righteous cynicism.
We are pleased to announce our latest work, in which we report the characterization of a new monoamine oxidase family member in vertebrates (MAO C). We collaborated with PhD students, postdocs, research assistants, and colleagues 😊 @USanSebastian biorxiv.org/content/10.110…
Brooke is the most talented,productive and empathetic scientist I personally know- if you are a young marine ecologist looking to join a group with a bright future and excellent mentorship, you should strongly consider joining to the Weigel lab at @HopkinsMarine @Stanford !
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”
Love this article by @schubertcm @geekwire on what we're trying to do here in #Seattle at @Xaira_Thera @JustasDauparas @naterbennett0 geekwire.com/2024/inside-th…
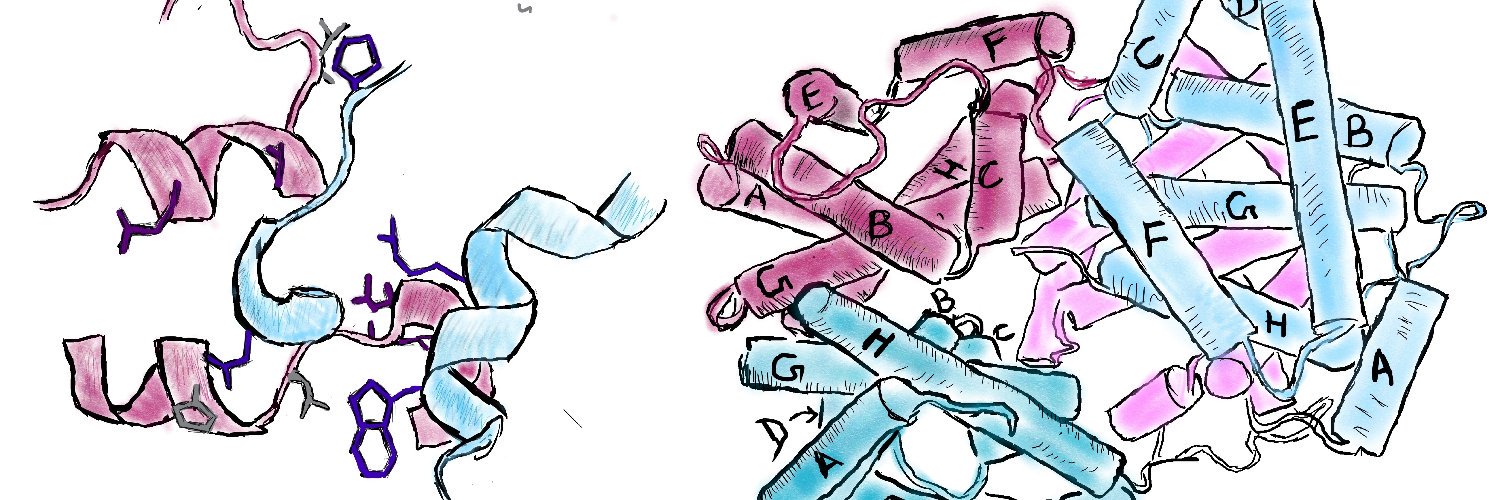
This is a monumental paper from Arvind. Not only did work out in his PhD how the classic model for allostery haemoglobin evolved. Now he designed allosteric proteins entirely from scratch. It think that's an awesome arc. nature.com/articles/s4158…
Awesome paper in @Nature! @ArvindSPillai1 achieved theoretically perfect ligand binding cooperativity with a hill coefficient of ~4 (hemoglobin is ~2.8 by comparison). Congrats Arvind, Abbas, @FloPraetorius and team, truly an honor to help with this work: nature.com/articles/s4158…
Super excited to share that our work on de novo design of allosteric proteins is now out in nature!👩🏽🔬 Huge thank you to @ArvindSPillai1 for his amazing mentorship and letting me be a part of this! nature.com/articles/s4158…