Jinwei Zhang Lab
@jinweizhang
RNA structural biology lab at NIH; believes #RNA lives and interacts in 3D space. All views my own.
Our @ScienceMagazine paper is out! Discovered how tRNA-derived small RNA (tsRNA/tDR) protects kidneys via RNA autophagy. science.org/doi/10.1126/sc… Huge thanks to @SaumyaDas_Lab and all our amazing collaborators! Excited for the therapeutic potential ahead! @MGHanesthesia @mghcvrc
I am so incredibly excited to share our latest work, on the exploration of #RNA secondary structure ensembles and discovery of RNA regulatory structural switches in bacteria and human cells, out today in @NatureBiotech: nature.com/articles/s4158…. A short tread! (1/n)
New paper by Kwok Lab @kitkwok6. RNA G-quadruplex structure targeting and imaging: recent advances and future directions. bit.ly/45C3AJf
New online! Structural basis for mTORC1 regulation by the CASTOR1–GATOR2 complex bit.ly/4lNDjwM
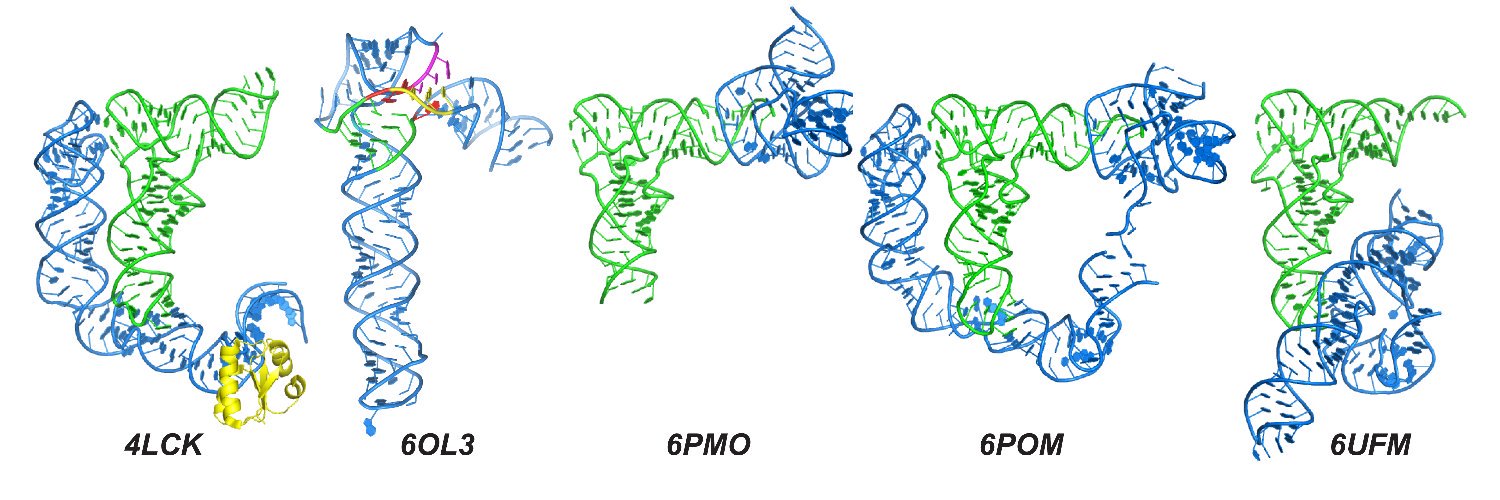
Pleased to share our new article on alternative protonation states of RNA: "Identification and characterization of shifted G•U wobble pairs resulting from alternative protonation of RNA" out in @NAR academic.oup.com/nar/article/53…
Out Now! Structure of an archaeal ribosome reveals a divergent active site and hibernation factor bit.ly/3UhiSfp #Archaea #RibosomeStructure #PeptidylTransferase
Our new paper is out in PNAS! We show that competition for binding to the Hfq matchmaker disfavors mismatched sRNA-mRNA combinations while allowing matched sRNA-mRNAs to form stable pairs. Kudos to Jorjethe Roca and Yi-Lan Chen for their fantastic work! doi.org/10.1073/pnas.2…
This week we publish in @NatureBiotech "High-precision cytosine base editors by evolving nucleic-acid-recognition hotspots in deaminase". In this work, we show that both nucleotide and context specificity of deaminases can be reprogrammed by hotspot-focused directed evolution 1/n
New paper out in Mol Cell, where we show how N-myristolytransferases lipidate proteins on the translating ribosome! Continuing our successful collaboration with @DeuerlingLab & Shu-ou Shan @Caltech @snsf_ch @ETH_en cell.com/molecular-cell…
A new #KwokLab paper has been released in Org. Biomol. Chem.: "G4-SLSELEX-Seq-driven discovery of a G4-specific targeting L-RNA aptamer with unique structural features." Congrats to the authors👏 doi.org/10.1039/D5OB00… @CityUHongKong @CityUChem @CityUScience @OrgBiomolChem
Discovery of a novel RNA polymerase III transcribed Alu RNA reveals a canonical and non-canonical tertiary structure and significant differences in association with the Alu RNA protein binding partner. bit.ly/3TFUrbf
Big congratulations to @KellyTHD_Nguyen 👍👍👍Super beautiful structure👍👍👍Very happy to contribute some RNA structure work to this great collaboration😊 @JohnInnesCentre @CellBiol_MRCLMB science.org/doi/10.1126/sc…
More than 2700 3′UTRs are highly conserved. These 3′UTRs are essential components in mRNA templates, as their deletion decreases protein activity without changing protein abundance. Highly conserved 3′UTRs help the folding of proteins with long IDRs. biorxiv.org/content/10.110…
With @ThirumalaiArmy and @tienhung_91, we wrote a review on 'Minimal Models for RNA Simulations', now published in @COSB_CRSB. We highlight how coarse-grained models capture the complex behaviour of RNA, from ribozyme folding to phase separation and more. doi.org/10.1016/j.sbi.…
New paper alert! Congratulations to Manju Ojha et al. We have shown how structural plasticity of RRE modulates nuclear export of HIV -1 RNAs. Mutation-driven RRE stem-loop II conformational change induces HIV-1 nuclear export dysfunction academic.oup.com/nar/article/53…
Mammalian tRNA acetylation determines translation efficiency and tRNA quality control doi.org/10.1038/s41467…
Now online! Pseudouridine RNA avoids immune detection through impaired endolysosomal processing and TLR engagement dlvr.it/TLfrWv
New @embojournal cover!🎉 The July issue features a stunning illustration by Dominika Dobosz (MCB) highlighting research by Biela et al. (@GlattSebastian group) on how #pseudouridine shapes human tRNAs—boosting stability for efficient protein synthesis. @JagiellonskiUni
New online! Structural basis for the synergistic assembly of the snRNA export complex bit.ly/44fJ1RP
@DjuranovicLab is from today officially part of @BrownUniversity and Brown RNA center. We will have our new website soon. rna.brown.edu