SheqLab
@ShechnerLab
The Shechner lab in UW Pharmacology. We study Noncoding RNAs and cellular architecture. He/His/Him. All comments are my own. @ShechnerLab.bsky.social
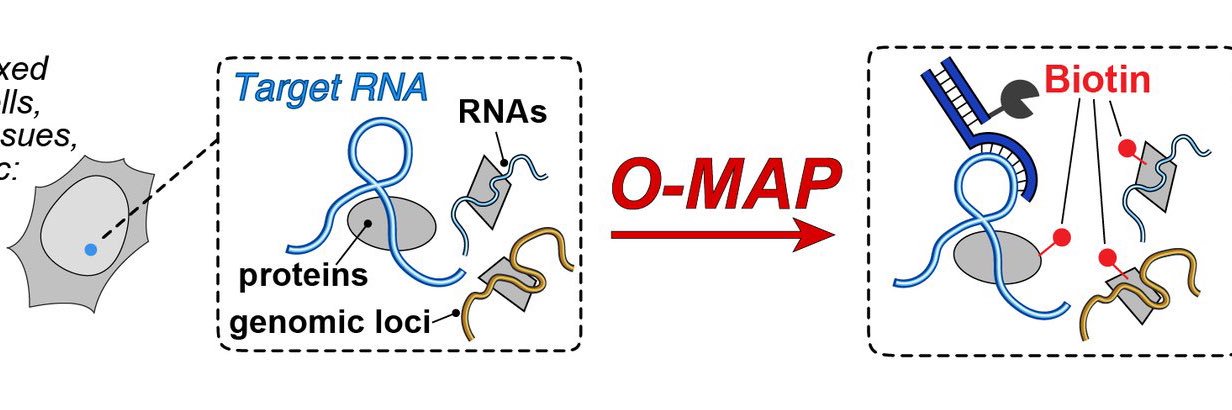
It's my pleasure to present the next big preprint from SheqLab! An exciting application of our O-MAP platform that I hope will transform the study of nuclear architecture. If you've ever wanted to dissect the subnuclear "neighborhood" around an individual locus, read on! (1/30)

Thank you so much for posting! And thanks to the inimitable @cellseywoodruff for the amazing writeup!!
#RNAsequencing helps map #RNA interactions with proteins and #DNA, revealing cellular processes and disease mechanisms. The O-MAP method by the @ShechnerLab enables precise RNA neighborhood mapping for deeper insights. - @UW rna-seqblog.com/o-mapping-the-…
*Huge* win for @genentech! Congrats, @DNAmyStrom!!
I'm thrilled to announce my next career step-- I’m joining Genentech as a Principal Scientist & Lab Head in Discovery Oncology! I’ll be hunting new ways to target cancers using my background in disordered nuclear proteins.
Amazing work, as always, from @Mayr_Christine’s lab. Congrats to all authors!!
More than 2700 3′UTRs are highly conserved. These 3′UTRs are essential components in mRNA templates, as their deletion decreases protein activity without changing protein abundance. Highly conserved 3′UTRs help the folding of proteins with long IDRs. biorxiv.org/content/10.110…
Awesome work from the Rouhanifard lab (as per usual 😊)
New from the Rouhanifard Lab! What if we could sort single cells based on whether a specific gene was actively being transcribed—and then ask what drives that burst? Introducing: NuclampFISH
Hello RNA World! Ever wonder what's "talking to" your favorite transcript, but were too scared to ask? In our review in @CellReports, @FKHM and I highlight new RNA-focused tools for discovering RNA interactions across organizational scales. Checkit! tinyurl.com/ydn6e3ac

🤩🤩🤩
Check out our new study on water networks in high resolution cryo-EM RNA data! @chiulab @RDasLab
TOP FIVE METABOLIC PATHWAYS …Based on whether they sound like the name of an old-school anime: 5. The Pentose-Phosphate Shunt 4. Peptidoglycan Degradation 3. Fatty Acid Beta-Oxidation 2. The Dark Reactions of the Calvin Cycle 1. Gluconeogenesis
Woohoo! So excited to see this important work out in print! Congrats @nandangokhale and all authors!
Our work on cellular RNA interactions with MAVS that promote antiviral signaling is finally live. So grateful to our indispensable collaborators as well as reviewers who helped to strengthen our work. #RNA #innateimmunity #virology @RamLabUW science.org/doi/10.1126/sc…
Woohoo! Congratulations @arjunrajlab for this well-deserved honor!!
Congratulations to @arjunrajlab, Professor in #PennBioengineering, who has been named the inaugural recipient of the Richard K. Lubin Professorship by @PennEngineers. Read more: bit.ly/4fCj4ik
Awesome work as always! Congrats to the whole team!!
Our latest preprint "Sequential structure probing of cotranscriptional RNA folding intermediates" is on bioRxiv. biorxiv.org/content/10.110… Led by @CourtneySzyjka and anchored by Skyler Kelly, this preprint describes a method for detecting cotranscriptional RNA folding transitions.
