
Josh Tycko
@JoshTycko
Postdoc at Harvard Med, Greenberg lab & BIDMC, Pollak lab aka DJ Fishing Expedition
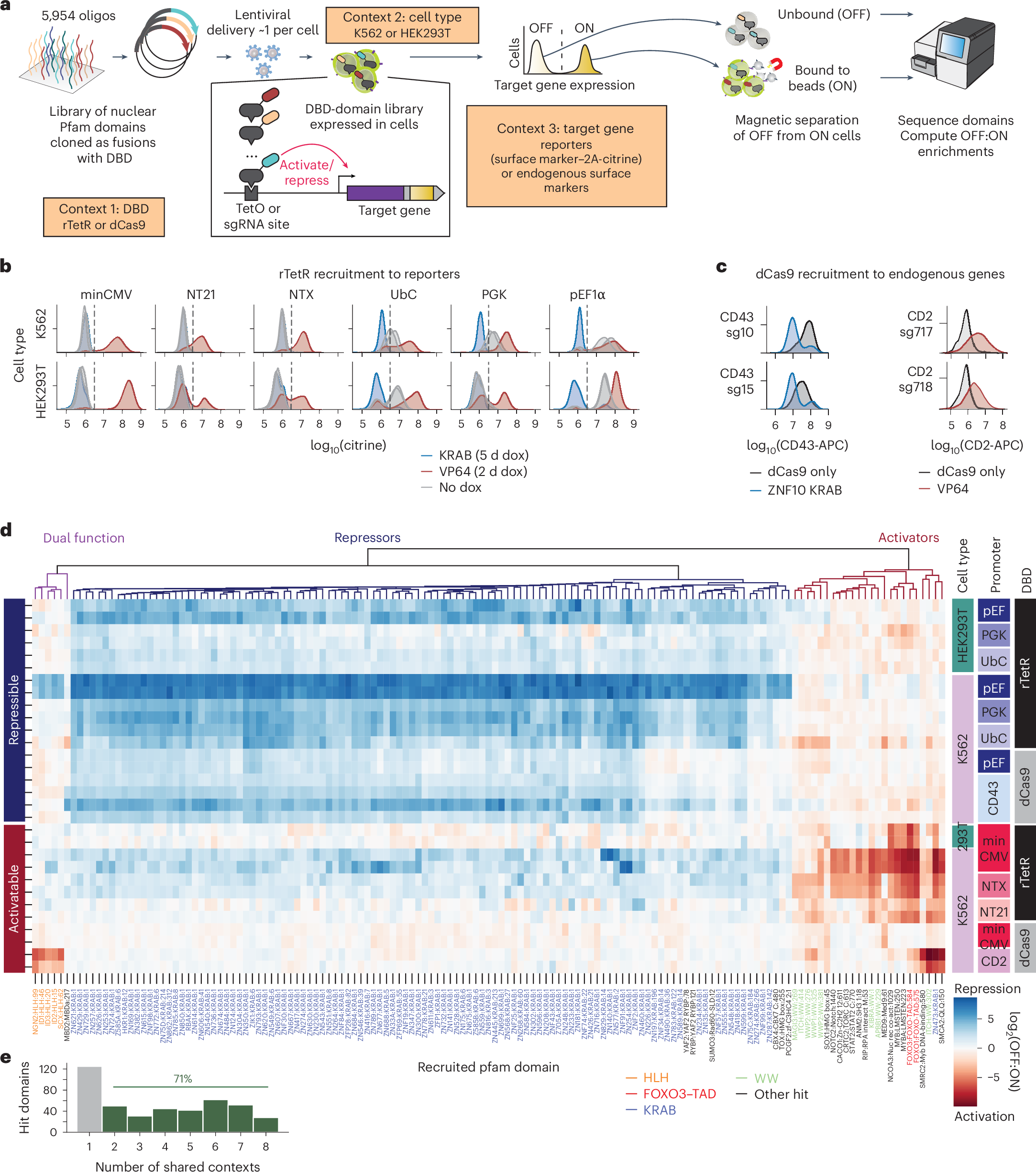
Our new paper on epigenome editing is out! We mapped which effector domains regulate transcription across genomic, cell type and DNA-binding domain contexts. Then we built new gene repressor and activator tools with improved efficiency & deliverability nature.com/articles/s4158…
thank you SPARK for supporting our work in its early days!
@stylus_medicine emerges from stealth to develop in vivo genetic medicines The company was co-founded by Stanford professors, who received funding and support from SPARK sparkmed.stanford.edu/stylus-medicin…
The machine-guided humanization paper is out after peer review! doi.org/10.1016/j.cels… Key contributions from current and previous international students on visas, representing 5+ countries.
So we beat on. Gene/cell therapies often use proteins that could be recognized/rejected as non-self by our immune system. We combine algorithms to build proteins that are therapeutically relevant and can masquerade as our own parts doi.org/10.1101/2025.0…
In a medical milestone, a customized base editor was developed, characterized in human and mouse cells, tested in mice, studied for safety in non-human primates, cleared by @US_FDA for clinical trial use, manufactured as a complex with an LNP, and dosed into a baby with a severe,…
Recombinases are simple short proteins that are sufficient to integrate long DNA with high specificity Combined with simple lipid nanoparticles, this could be a way to make durable cell therapies affordable and distributable
Stylus originated from a wonderful scientific collaboration discovering serine DNA recombinases with Ami Bhatt, @BintuLacra, and @MCBassik spearheaded by star researchers @mgdurrant @AlisonFanton @JoshTycko A huge thanks also to Cosmas Giallourakis and @eminkow for their…
So excited for the Stylus team and their progress turning recombinases into therapeutics!
Delighted to publicly launch @Stylus_Medicine today to program the human body into a cell therapy factory to fight cancer. It's a privilege to work with Emile and Jason as CEO/CSO of Stylus and our earliest believers Josh Resnick/Ness Bermingham from RA Capital + @khoslaventures
How Multiplexed Gene Fragments are Expanding What’s Possible in Molecular Biology! @TwistBioscience @JoshTycko Moving Beyond 300 Base Pairs | Twist Bioscience buff.ly/JZ8Ug9E
Check out this new preprint showing how to engineer proteins for therapeutics while mitigating immunogenicity!
So we beat on. Gene/cell therapies often use proteins that could be recognized/rejected as non-self by our immune system. We combine algorithms to build proteins that are therapeutically relevant and can masquerade as our own parts doi.org/10.1101/2025.0…
Joining the culture war! We tried liquid culture to re-amplify a lenti sgRNA plasmid library from @Addgene. N=1 but it wasn't worse than our traditional large plate protocol, and seems even better in uniformity and liquid culture is easier to do thanks @nmateyko @CarldeBoerPhD
Do you really need to spread plasmid libraries on mountains of plates to get uniform growth, or can you just dump them in a flask and call it a day? We make huge plasmid libraries in the de Boer Lab, so we tested whether culture method really matters. 1/ biorxiv.org/content/10.110…
Today we report that an engineered skin bacterium, swabbed gently on the head of a mouse, can unleash a potent antibody response against a pathogen. Could lead to topical vaccines that are applied in a cream. @DjenetBousbaine led the charge... @Nature 1/55
A dream come true: in 2004 I started thinking about counting microstates to predict gene expression with @JKondev Rob Phillips @HernanGGarcia. 20 years later we can measure them: nature.com/articles/s4158… Thank you @WJGreenleaf @bgrdoughty @MichaelaThinks Julia and all co-authors!
Incredibly excited that our paper linking single molecule states of TF binding to gene expression using quantitative thermodynamic models is out in Nature today. An amazing collaboration with the Bintu Lab. Congrats to Ben, Michaela, and Julia! nature.com/articles/s4158…
🧬 Thrilled to share our latest work on engineering precise DNA recombinases for targeted genome integration! Through directed evolution & protein fusions, we achieved over 50% efficiency & 97% specificity for multi-kb DNA cargo insertions. Big thanks to all co-authors -
🧵In new work, we report a systematic engineering roadmap to optimize large serine recombinases (LSRs) for direct, site-specific insertion into the human genome 🧬. We achieved over 50% insertion efficiency and 97% genome-wide specificity, a 10X improvement over our previous work
Development of compact transcriptional effectors using high-throughput measurements in diverse contexts go.nature.com/3NMu00N