Xiaojing Gao
@SynBioGaoLab
Assistant Professor @ Stanford ChemE synthetic biology, biomolecular engineering https://synbiogaolab.bsky.social/
Can we use RNA to insert kilobases of DNA into the human genome, opening up a new way to engineer cells or treat disease? Excited to share our latest work turning a retrotransposon (a “jumping gene” with an RNA intermediate) from songbirds into a genome editing tool. 🧵 (1/7)
The Blum lab is recruiting graduate students and postdocs to work on projects focused on food allergy and oral tolerance! Join our new group at the Salk Institute! Grad students: Reach out to me if you're interested in a rotation! Postdoc candidates: recruiting2.ultipro.com/SAL1013SIBS/Jo…
🚀 Join CongLab @Stanford! We’re hiring postdocs to create lab-in-the-loop self-evolving AI agents, open benchmarks, to design, test & learn—advancing safer gene & cell therapies. Build on CRISPR-GPT, RNAGenesis model, Genome-Bench, for innovative medicines. #PostdocJobs
We're hiring @kemijski ! We aim to translate technologies of synthetic biology toward therapeutic applications, using both protein and RNA-based strategies. PhDs or MSc excited about #syntheticbiology #biotech #proteindesign #RNAdesign are invited to inquire and apply. Pls RT🔁
🚨 New Preprint 🚨 A continuous landscape of signaling encodes a corresponding landscape of CAR T cell phenotype How can we tune JAK/STAT signaling to create better immune cell therapies? doi.org/10.1101/2025.0…
Synthetic biology could enable new types of programmable therapeutics. Our new preprint introduces synthetic protein circuits that selectively trigger cell death in Ras-mutant cancer cells, with interesting advantages compared to existing approaches. biorxiv.org/content/10.110…
The machine-guided humanization paper is out after peer review! doi.org/10.1016/j.cels… Key contributions from current and previous international students on visas, representing 5+ countries.
So we beat on. Gene/cell therapies often use proteins that could be recognized/rejected as non-self by our immune system. We combine algorithms to build proteins that are therapeutically relevant and can masquerade as our own parts doi.org/10.1101/2025.0…
New preprint! We carried out the largest complementation and inhibition study for diverse homologs in a single protein family (DHFR) to date. Highlights thread follows... 1/n 🧵
Exploring Antibiotic Resistance in Diverse Homologs of the Dihydrofolate Reductase Protein Family through Broad Mutational Scanning biorxiv.org/cgi/content/sh… #biorxiv_synbio
Plasmid-based reporter assays are the bedrock of regulatory genomics. But a basic question has gone unanswered for decades: Do chromatin architectures form on plasmids transfected into mammalian cells—and does it matter? We finally have answers. biorxiv.org/content/10.110…
AASF cautions harm to US talent pipeline from new State Dept policy to aggressively revoke visas for Chinese students, including in critical fields. "It sends a chilling message to the world that America no longer welcomes global talent" - @GKusakawa aasforum.org/2025/05/29/asi…
nature.com/articles/s4158… Awesome new paper from Shan Za and @ColumbiaBiochem and systems biology colleague Chaolin Zhang @chaolinzhang
Did you miss the abstract submission deadline for #mSBW2025? We have a few spots open, but you must submit by tonight (May 12) at midnight, ET. The sessions will cover cutting-edge developments and offer a dynamic forum for discussion and collaboration. bit.ly/3WwfChK
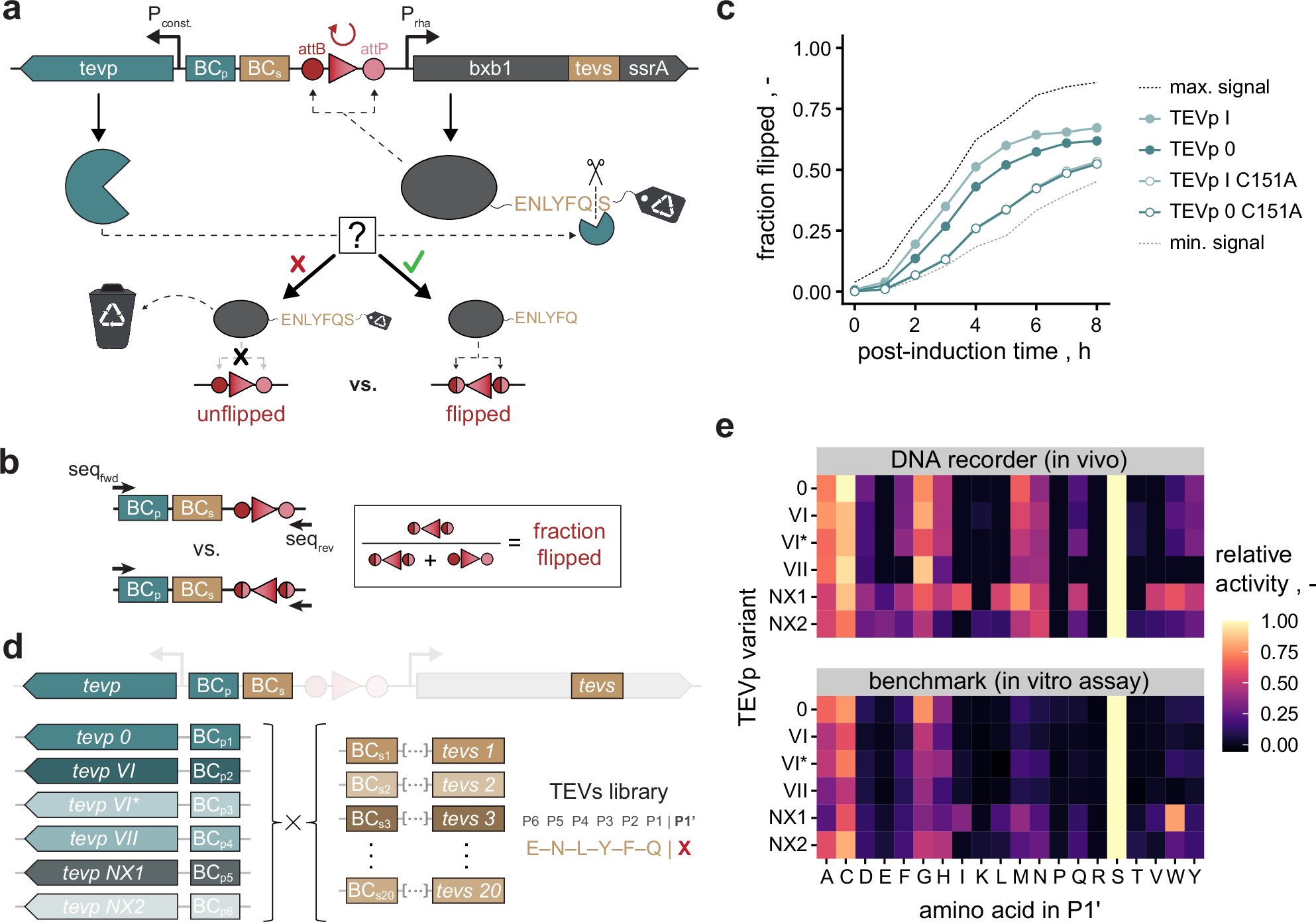
Our new paper is just online. Congratulations to Junjie and the team.Super Recombinator (SuRe): An in vivo recombination system for scalable and efficient transgene assembly at a single genomic locus | bioRxiv biorxiv.org/content/10.110…
Splitting proteins into two segments that reconstitute protein function by proximity is a great #synbio tool for recognition of proteins, cells and even regulation of therapy. BUT it requires two proteins, so twice the work and cost. Could it be done with just a single protein?🧵
Editing 14 genes is no small feat, but I am not seeing enough evidence that "it is a dire wolf". Our community needs to communicate better to the public, now mor than ever, and imo that includes not exaggerating what we can deliver.
TIME's new cover: The dire wolf is back after over 10,000 years. Here's what that means for other extinct species ti.me/4jlJB54
Congrats to @ChappellLab (a @NUSynBio alumni!) on their latest in @NatureBiotech : nature.com/articles/s4158… Amazing RNA engineering applied to track microbial ecosystems!