Dhiraj Indana
@DhirajIndana
@DamonRunyon @HHMINEWS Postdoc @Elowitzlab | Previously, PhD @theChaudhurilab, BTech @iitroorkee
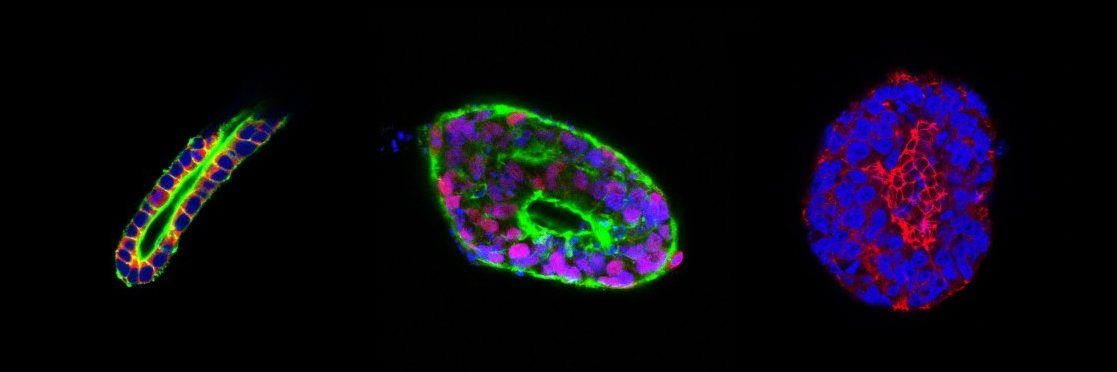
📢 Incredibly excited to share the peer-reviewed version of our lumenogenesis work. We developed a human epiblast model and discovered a new mechanism of lumen expansion that is driven by actin! @CellStemCell @theChaudhurilab 🧵 [1/17] doi.org/10.1016/j.stem…
![DhirajIndana's tweet image. 📢 Incredibly excited to share the peer-reviewed version of our lumenogenesis work. We developed a human epiblast model and discovered a new mechanism of lumen expansion that is driven by actin! @CellStemCell @theChaudhurilab 🧵 [1/17]
doi.org/10.1016/j.stem…](https://pbs.twimg.com/media/GM_ZkE7aQAA0_Uw.jpg)
Our logo is a cell reaching out 🧫, a figure in motion 🏃♂️, and a network of signals 🌐! It captures our research: Mechanism (Discovering principles of life), Function (Elucidating the roles of living systems), and Design (Building synthetic circuits). bit.ly/ShiyuXiaLab
If you’ve been following this account closely, you might already know methods to probe mechanics in vitro, but what about in live embryos? I’m Arthur Michaut , and I’m going to share a few great papers on this aspect of #EpithelialMechanics.
Have you always wanted to take a protein from its native context and make it work elsewhere? Our novel sampler computationally “cytosolize” a secreted enzyme while maintaining its structure, generalizable to other multi-objective guided generation tasks biorxiv.org/content/10.110…
In January 2026, I will be joining Berkeley @UCBerkeley as an Assistant Professor in the Department of Chemical and Biomolecular Engineering, College of Chemistry @UCB_Chemistry, with exciting connections with world-class communities including the Lawrence Berkeley National…
Have you ever thought about inflating tissues? Or maybe quickly deflating those inflated tissues? New #EpithelialMechanics pre-print: doi.org/10.1101/2025.0… 🧵with pressure control, multiscale buckling, patterned wrinkling
🚨 Second preprint of the week! We uncover the multiscale dynamics of active viscoelastic buckling in epithelia. We harness these mechanical instabilities for synthetic morphogenesis. Led by @onenimesa🙌. Theory by Marino Arroyo and team. biorxiv.org/content/10.110…
How to turn a layer of fibroblasts into a tulip 🌷? Check out our new pre-print on shape-programmable living surfaces. Led by @pauguillamat @IBECBarcelona biorxiv.org/content/10.110…
🚨 New Preprint 🚨 A continuous landscape of signaling encodes a corresponding landscape of CAR T cell phenotype How can we tune JAK/STAT signaling to create better immune cell therapies? doi.org/10.1101/2025.0…
Synthetic biology could enable new types of programmable therapeutics. Our new preprint introduces synthetic protein circuits that selectively trigger cell death in Ras-mutant cancer cells, with interesting advantages compared to existing approaches. biorxiv.org/content/10.110…
🚨📢Fresh off the press and featuring new exciting experiments! 🧪 We show how glycolytic activity instructs germ layer proportions through regulation of Nodal and Wnt signaling doi.org/10.1016/j.stem… B2B with @VillarongaAlba @teamstembryo : doi.org/10.1016/j.stem…
Can metabolic processes instruct cell fate decision making during embryonic development? In our new preprint, we show an important role of glycolytic activity for the development of endoderm and mesoderm through the regulation of Nodal and Wnt signalling. biorxiv.org/content/10.110…
Excited to share our new paper out in @Nature revealing cell-type specific nuclear organization and its link to gene regulation using new spatial multi-omics technologies! nature.com/articles/s4158…
Very excited this work is out in @NaturePhysics. A wonderful collaboration between my group @MIT and @PhysOfLifeLMU.
Optogenetically induced chemo-mechanical excitations are used to drive and study shape deformations in starfish oocytes. Understanding and eventually controlling such waves is important for the development of synthetic cells. nature.com/articles/s4156…
LIDAR is out! rdcu.be/efugD We've improved a lot, e.g., making MESA-like LIDAR work, RNAseq of on-/off-target editing, and optimizing RNA delivery. We are one baby step closer to making it useful for therapeutics. NIH high-risk high-reward program provides key support.
Following our field's inspiring history of building synthetic receptors, here we share our lab's first attempt at a post-transcriptional one. Based on ADAR-editing and compatible with various inputs/outputs.
New preprint countdown! Mitochondria are cells within our cells. They need the same core activities - replication, transcription, translation. How do cells enable these diverse activities in both compartments? We uncover an unexpected strategy with ancient origins. Stay tuned!
Theory predicted two kinds of fibrosis, hot and cold. We show in vivo that hot and cold fibrosis describe heart failure, and heart attack, respectively, showing that they are very different disease states. @MiyaraShoval @MiriAdler @Tzahore
Our work made it to the cover of @CellSystemsCP! 🥳 Cold and hot fibrosis define clinically distinct cardiac pathologies Check out our paper here: cell.com/cell-systems/f… @UriAlonWeizmann @Tzahore @MiriAdler @WeizmannScience @HebrewU
Stoked to present our latest, brilliantly led by Chris et al & Alejandro Torres-Sanchez. How tissues are patterned during development – we found that geometry-constrained ECM fractures pattern the myocardium in the vertebrate heart 1/n biorxiv.org/content/10.110…
Excited to contribute to the pioneering work at @DamonRunyon @HHMINEWS @ElowitzLab!
We are delighted to announce our most recent class of Damon Runyon Fellows! These 13 scientists will receive four years of independent funding to investigate cancer causes, mechanisms, therapies, and prevention. Read more: bit.ly/4kv5JLF
Our lab doors opened on Monday @ucsdbe and we had lab lunch today! Very excited to have three grad students (Sophia, Ruturaj, and Xiaoya) and one undergrad (Josie) rotating. More of our scientific adventures to come😎!
So we beat on. Gene/cell therapies often use proteins that could be recognized/rejected as non-self by our immune system. We combine algorithms to build proteins that are therapeutically relevant and can masquerade as our own parts doi.org/10.1101/2025.0…
Announcing Evo 2: The largest publicly available, AI model for biology to date, capable of understanding and designing genetic code across all three domains of life. arcinstitute.org/manuscripts/Ev…