
Gang Fang
@gangfang_1
Long read (DNA, RNA) | Epigenetics (bacteria, human) | Professor of Genomics @IcahnMountSinai | Humid Lab🧫💻🧪 https://bsky.app/profile/gangfang.bsky.social
Glad to share our preprint 🏃**LongTrack**: long read metagenomics-based precise tracking of bacterial strains (and their genomic changes!) after fecal microbiota transplantation. A 4+ years journey and tour de force by ** @fanyu48696214 ** and team. A long 🧵 💩Fecal microbiota…

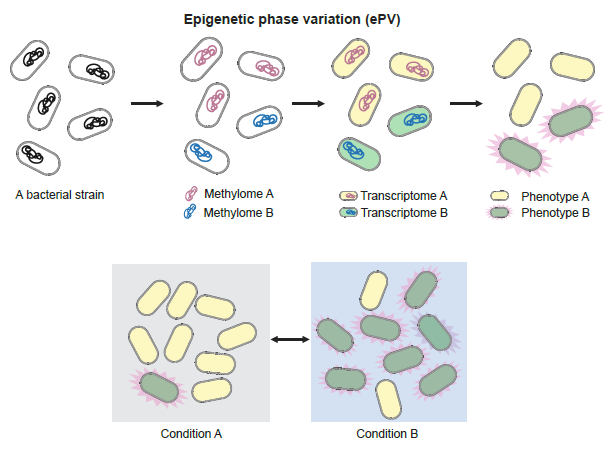
Thrilled to share our new preprint: Epigenetic phase variation in the gut microbiome enhances bacterial adaptation 🔎a hidden layer shaping our gut microbiome. A 5+ Year journey by @Fannimi2001 👏 and team @fanyu48696214 @MagdalenaKsi, incredible collaboration with labs of…
Happy to share our latest work published in @CD_AACR led by Mei Luo @JingwenYang_! Great collaboration with @NCIEytanRuppin Gordon Mills! @IUCancerCenter @HealthIusm60944 @iubioinfo
Now online in @CD_AACR: Ancestral Differences in Anticancer Treatment Efficacy & Their Underlying Genomic & Molecular Alterations - by Mei Luo, @JingwenYang_, Gordon Mills, @NCIEytanRuppin, @lenghan_bioinfo, et al doi.org/10.1158/2159-8… @OHSUKnight @theNCI @IUCancerCenter @IUPUI
Amazing work by Dr. Rechkoblit et al. from the Aggarwal and @gangfang_1 Labs on CBASS Cap5 endonuclease showing bacterial self-destruction by digesting its own DNA! Congratulations! 👏 #bacteria #cbass #cap5 #proteinstructure #immunology #virology nature.com/articles/s4146…
argNorm is published now academic.oup.com/bioinformatics…
HEY HEY #AMR people, the argNorm preprint is now available on @QUT ePrints: eprints.qut.edu.au/252448 We* designed a tool for normalizing ARG annotations across currently popular tools and databases. * @svetlana_up @VedanthRamji @HuiChong6 @cocodyq @FinlayM @luispedrocoelho 1/6
Interested in #LongReadSequencing? We are delighted that @GangFang_1 (Professor @IcahnMountSinai) is joining us at #FOGBoston, sharing insights on detecting DNA modifications using long read sequencing. Register now: hubs.la/Q03glPF80
I’m so proud and beyond thrilled to share the final product of my thesis @SinaiBrain now published @Nature! Here, we explored the complex functional impact of rare neurexin-1 deletions across human glutamatergic and GABAergic neurons! nature.com/articles/s4158…
1/ Excited to share our new study in @CellCellPress :"Conserved genetic basis for microbial colonization of the gut" We present a tree-of-life-scale genotype-habitat association that reveals conserved gene modules in gut microbes—and identifies novel colonization factors.
Computational discovery and experimental validation of mammalian gut colonization factors. Work led by our own amazing @menghan_liu in collaboration with @SohailTavazoie lab at @RockefellerUniv. @Columbia_Bio @ColumbiaSysBio @ColumbiaBiochem cell.com/cell/fulltext/…
Computational discovery and experimental validation of mammalian gut colonization factors. Work led by our own amazing @menghan_liu in collaboration with @SohailTavazoie lab at @RockefellerUniv. @Columbia_Bio @ColumbiaSysBio @ColumbiaBiochem cell.com/cell/fulltext/…
I'm honored to be inducted as a fellow into the American Institute for Medical and Biological Engineering @aimbe. Had a great visit to Capitol Hill and advocated for biomedical engineering research. @NU_BMG_SQE @NUFeinbergMed
Excited to share another collaborative study with Dr. Joe Costello - Intratumor heterogeneity of 3D genome organization in primary glioblastoma samples. Led by Qixuan and Juan, and congrats to the whole team! @LurieCancer @NUFeinbergMed @NeurosurgUCSF science.org/doi/10.1126/sc…
My lab at @GeiselMed has an open postdoc position in fungal metabolomics. We will combine isotope tracing, mass spectrometry data analysis, and mathematical modeling to understand metabolic rewiring in fungal cells. Research question depends on your interest. Please RT and apply!
Very interesting analogy to describe our LongTrack preprint on long read based FMT strain tracking! Thank you!
Tracking bacterial ‘seeds’ in new soil reveals how they grow, adapt, and pave the way for next-gen microbiome therapies. @gangfang_1 @fanyu48696214 @MagdalenaKsi @jeremiahfaith1 @grinsa01 @NOKaakoush @IcahnMountSinai @biorxivpreprint biorxiv.org/content/10.110…
Happy to share our @MolTherapy review on long-read RNA sequencing for exploring transcriptome complexity in human diseases, co-first-authored by our PhD students Isabelle Heifetz Ament @PennBiology and Nicole DeBruyne @CAMBUpenn! cell.com/molecular-ther…
One small step for X-inactivation, one giant leap for @theNestorLab. So happy that our first paper on XCI is out in @NatureComms. A study into human T-cell development took a wild turn when we found one female was fully skewed for XCI! nature.com/articles/s4146…
Out now in @Nature: Gut microbiota strain richness is species specific and affects engraftment @achenliaw @jeremiahfaith1 nature.com/articles/s4158…
Blood–brain barrier-crossing conjugates (BCC). Exciting collaboration with the teams @EricJNestler @aw_charney @Ramon_Parsons # Xiaolin Cheng! Thanks @NatureBiotech editors and reviewers, @IcahnMountSinai @IcahnInstitute @SinaiImmunol @SinaiBrain @TischCancer @BMEIIsinai
Intravenous administration of blood–brain barrier-crossing conjugates facilitate biomacromolecule transport into central nervous system go.nature.com/412exBi