
Devansh Pandey
@devpandeyay
PhD Candidate @UTexasILS | Biotechnology and Biochemical Engineering @IITkgp 2022 | Interested in Human Genetics, Evolution, and Deep Learning
Thrilled to share that the first project from my PhD is published in Nature Communications (@NatureComms) ! 🎉 We explored Holocene European aDNA to uncover how selective sweeps have been obscured by neutral processes like drift and admixture. Read more: nature.com/articles/s4146…

nature.com/articles/s4225… Another paper that produces a GWAS for unsupervised imaging phenotypes but my main issue is interpretability. What do I do with a PRS for latent vector 14 of the heart, or it's effect size? If you care about disease prediction, don't need IDPs at all
Our new paper on ancient DNA to address the spread of Uralic languages now out in @nature (we started working on this in 2018). Many interesting results, including a sample from ~16k years ago Yakutia that can be modeled as an unadmixed Native American. nature.com/articles/s4158…
Excited to share our new study just published in @ScienceMagazine, led by @xliaoyi! Check it out.
Thrilled to share that the first project from my PhD is now published in Science (@ScienceMagazine)! 🎉 We studied the genetic architecture of and evolutionary constraints on the human pelvic form. Read more: science.org/doi/10.1126/sc…
In a new paper, colleagues and I raise concerns about the conclusions and data from Zhang et al 2022, a paper which reported that they obtained DNA from a 14-thousand-year-old individual from Red Deer Cave in Yunnan China referred to as ‘Mengzi Ren’ (MZR) (Zhang et al Fig 1) 1/15
on twitter, you will always find someone who is tweeting your thoughts but with better grammar.
When in evolutionary time does accelerated genomic change occur across phenotypes? Newly published today in @CellGenomics , my latest manuscript with @mashaals and @vagheesh integrates comparative, functional, and ancient genomic data with GWAS. Read more: cell.com/cell-genomics/…
New paper led by star PhD student @mrschwob (who is currently on the statistics job market) building a bayesian hierarchical model to understand gene flow across space and time in ancient DNA datasets. academic.oup.com/biometrics/art…
Scientists from @UTAustin & @UCLA used ancient DNA samples to look back in time & reveal new insights into how ancient Europeans adapted to their environments over 7,000 years of European history. #AncientDNA @UT_Stats @Texas_IB @Vagheesh @DevPandeya cns.utexas.edu/news/research/…
Happy to share our recent publication in the Journal of Hydrology. Our study examines water and carbon fluxes from a supra-permafrost aquifer into Imnavait Creek, a headwater stream in Alaska’s continuous permafrost region. Link to paper: doi.org/10.1016/j.jhyd…
Delighted to share that our paper on discovering selective sweeps with haplotype statistics in aDNA is now out in Nat. Comm! Wonderful collaboration with @vagheesh, @devpandeyay, and @MarianaHarr.
Thrilled to share that the first project from my PhD is published in Nature Communications (@NatureComms) ! 🎉 We explored Holocene European aDNA to uncover how selective sweeps have been obscured by neutral processes like drift and admixture. Read more: nature.com/articles/s4146…
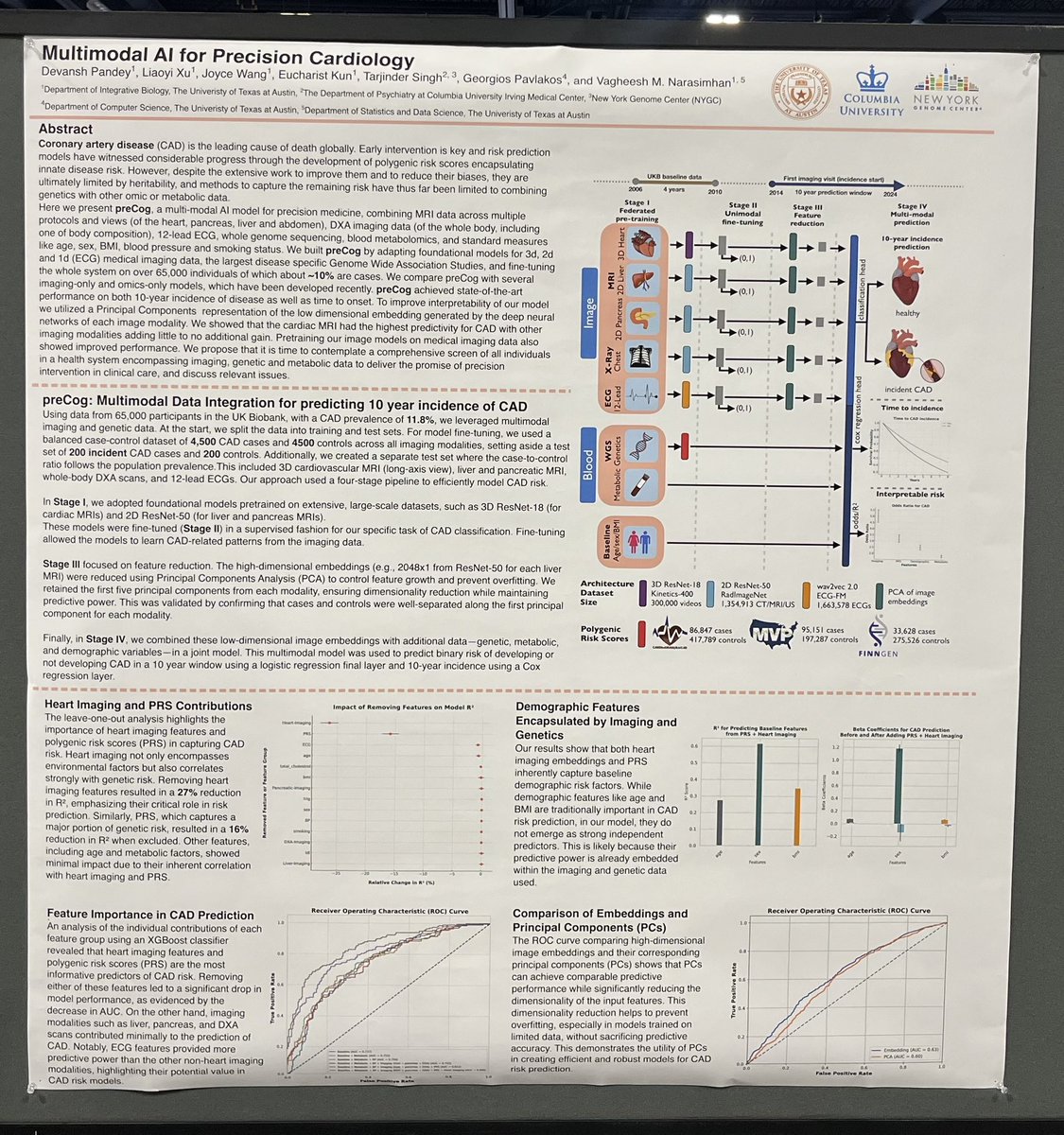
Exploring #MultimodalAI for disease diagnosis? Stop by my poster (5128W) today at #ASHG2024 from 2:30-4:30 pm – excited to share insights and connect!
Stoked to see our Bits to Binders protein design competition covered in @Nature News today alongside @adaptyvbio, @EvoscaleAI, and @Align_Bio! We just wrapped up the design stage of our comp, stay tuned for a update in the coming days! nature.com/articles/d4158…
Excited to share my 1st-author work from the @arbelharpak Lab! We examine the accuracy of polygenic score (PGS) predictions across individuals in the UK Biobank. We make 3 observations that expose gaps in our understanding of PGS “portability”. biorxiv.org/content/10.110… (1/28)
Announcing the BioML Challenge 2024: Bits to Binders! In this 5-week competition, teams will use AI methods to design protein binders that bind to a target antigen and activate an immune response from its associated CAR-T cell. Real-world scenario. With wet lab validation!
Check out this amazing hackathon by BioML society at UT Austin. It involves use of AI for protein design followed up by experimental validations.
Announcing the BioML Challenge 2024: Bits to Binders! In this 5-week competition, teams will use AI methods to design protein binders that bind to a target antigen and activate an immune response from its associated CAR-T cell. Real-world scenario. With wet lab validation.
Overall saw some great talks, met awesome folks in the #SMBE community, and enjoyed the food and culture of Puerto Vallarta. Thanks to all the SMBE organizers for their work and looking forward to the next one!