
Eucharist Kun
@EucharistKun
Working on the intersection of artificial intelligence and large-scale human genomics. Interested in all things cancer research related
Excited to announce that our manuscript is finally out and featured on the cover of @ScienceMagazine! This was a huge collaborative effort across multiple universities and I'd like to thank all who contributed. Check out our work here: science.org/doi/10.1126/sc… @TexasScience
Humans are the only bipedal great apes, due to the evolution of our unique skeletal form. What genomic regions underlie such change? Our cover article in @sciencemagazine led by @EucharistKun and in collab. with @tarjs is now out! science.org/doi/10.1126/sc…
Our paper led by @FlynnBrie on using AI as a phenotyping engine for diseases which are diagnosed largely through imaging is now out in @npjDigitalMed. We double case counts + obtain a quantitive biomarker associated with severity directly from imaging. nature.com/articles/s4174…
Thrilled to share that the first project from my PhD is now published in Science (@ScienceMagazine)! 🎉 We studied the genetic architecture of and evolutionary constraints on the human pelvic form. Read more: science.org/doi/10.1126/sc…
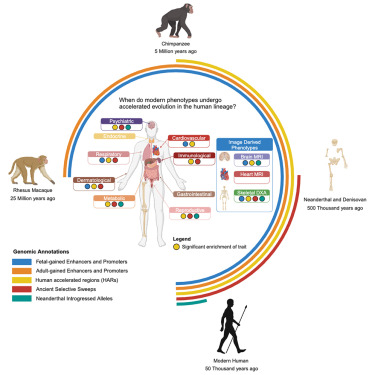
Tracing human trait evolution through integrative genomics and temporal annotations, a preview of our recent study with @EucharistKun and @vagheesh cell.com/cell-genomics/… @CellGenomics
Excited to share our manuscript “The trait-specific timing of accelerated genomic change in the human lineage” now out in Cell Genomics cell.com/cell-genomics/… with @EucharistKun and @vagheesh 1/n
When in evolutionary time does accelerated genomic change occur across phenotypes? Newly published today in @CellGenomics , my latest manuscript with @mashaals and @vagheesh integrates comparative, functional, and ancient genomic data with GWAS. Read more: cell.com/cell-genomics/…
When in evolutionary time does accelerated genomic change occur across phenotypes? Newly published today in @CellGenomics , my latest manuscript with @mashaals and @vagheesh integrates comparative, functional, and ancient genomic data with GWAS. Read more: cell.com/cell-genomics/…
Talks from @valenomics and @EucharistKun at #SMBE2024. Fun collaborations with @dortega_delv @emiliahsc and @vagheesh using ancient DNA, MXB, comparative genomics resources and multitudes of GWAS to ask questions about complex trait and disease evolution
A new release of the HARE pipeline for enrichment analysis (with lots of new functionality!) is live and published in @JOSS_TheOJ joss.theoj.org/papers/10.2110…. Cannot thank @EucharistKun and @vagheesh enough for all their work making v1.2 happen.
The obstetrical dilemma - a 60yr old hypothesis in evolutionary theory, suggests that a narrow pelvis aids bipedal locomotion but complicates childbirth. But how does it stand up to modern empirical evidence? Read our preprint to find out! biorxiv.org/content/10.110…
I’ll be presenting my work on leveraging genomic data for evolutionary analysis at 2:15 PM in the poster hall, come by and say hi! #ASHG23
Hey Y’all! I am presenting my work on LLMs for genotype imputation at #ASHG23. Drop by my poster and we’ll talk about the interesting science behind our work.
Excited for #ASHG23 with the rest of the Narasimhan lab! Featuring @xliaoyi’s platform talk on “The genetic architecture and consequences of the human pelvic form” during Session 021 as well as posters from @devpandeyay, Joyce Wang, and myself. Come see what our lab is up to!

In this week's #SciencePodcast🎙️ with @boron110, @sandeeprtweets, and @EucharistKun: ▶️ Adding thousands of languages to the #AI lexicon ▶️ The genes behind our bones 🎧 Listen here: scim.ag/3r4
Genome-wide map reveals regions associated with skeletal changes that enabled humans to walk upright go.nature.com/3K9zb9v
First time on a podcast and it was a great experience, thanks for having me! @boron110
In this week's episode of the @ScienceMagazine Podcast: Adding thousands of languages to the AI lexicon w/ @sandeeprtweets, and the genes behind our bones w/ @EucharistKun science.org/doi/10.1126/sc…
What you can get when you combine deep learning #AI of DXA scans and genomics in >30,000 people! @ScienceMagazine @uk_biobank @ScienceVisuals doi.org/10.1126/scienc… science.org/toc/science/cu…
World-changing work from @TexasScience! Our evolutionary scholars just landed on the cover of @ScienceMagazine for research that could help doctors better predict patients’ risks! 🤘 #WhatStartsHere
By combining data from full-body x-ray images and associated genomic data from more than 30,000 UK Biobank participants, researchers have provided new insights into the evolution of the human skeletal form. Learn more this week in Science: scim.ag/3qw