Jinghui
@Jinghui_Song
Supervisor @genomics212 | Former Postdoc @KunZhangBioE lab | Former PhD @YiChengqi lab | Single-cell genomics | Epitranscriptomics | Spatial Transcriptomics
So excited to share our work (cell.com/molecular-cell…). We developed a programmable RNA base editor, termed RESTART in the lab of Prof. Yi (@YiChengqi). Hoping our work will be a promising tool for therapeutics of PTC-disease. cell.com/molecular-cell…
‘Jumping gene’ may have erased tails in humans and other apes—and boosted our risk of birth defects science.org/content/articl…

Our enhanced version of RNA base editor RESTART is online now! RESTART v3 utilizes near-cognate tRNAs to improve the readthrough efficiency of pseudouridine-modified PTCs. The talented work from Yi Lab (@YiChengqi). nature.com/articles/s4158…
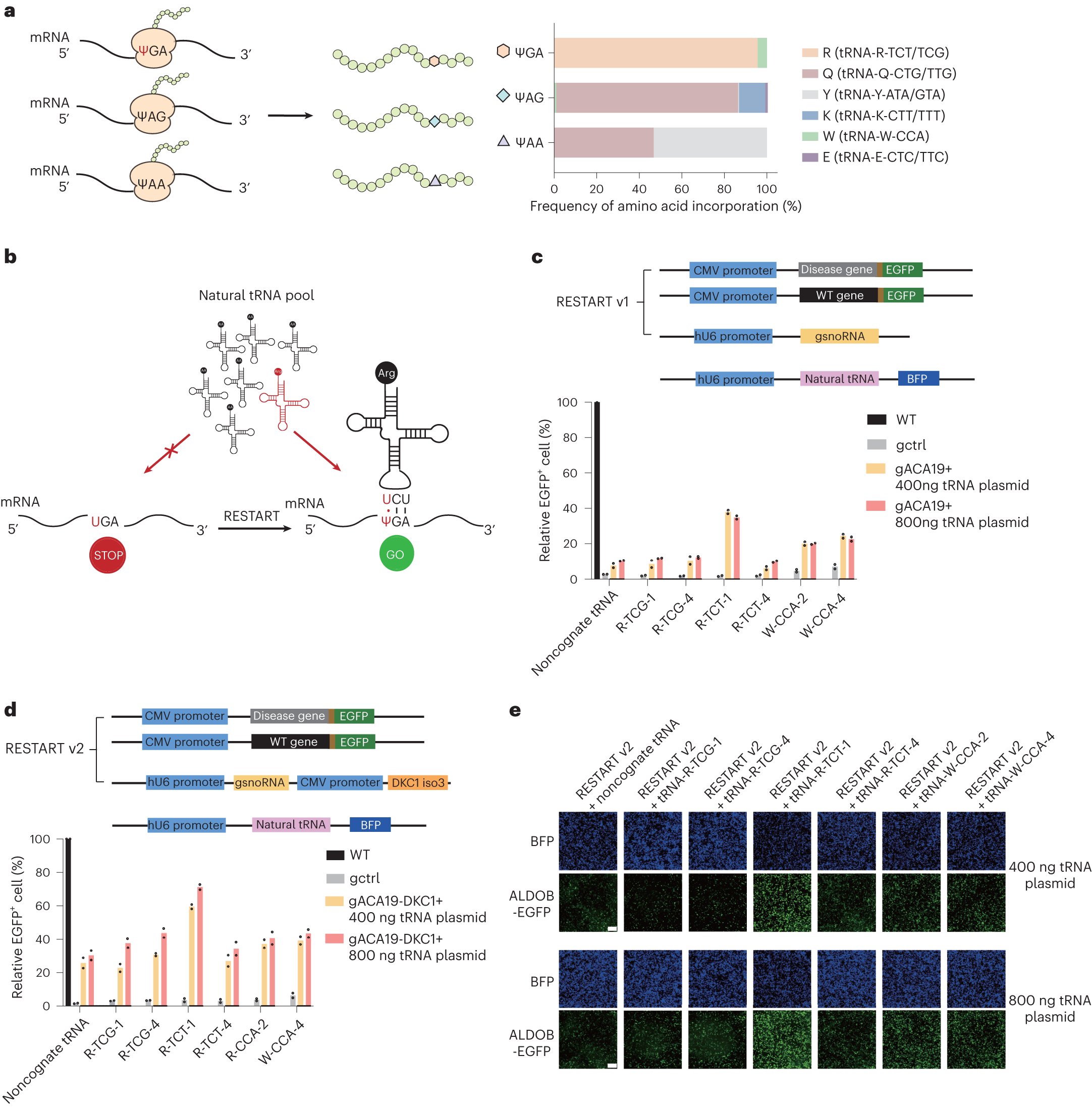
Programmable RNA base editing via targeted modifications | Nature Chemical Biology nature.com/articles/s4158…
Honored that our RNA base editor "RESTART" has been selected as the best technology of Molecular Cell 2023. Huge thanks to my mentor Prof. Yi (@YiChengqi). …bestof2023.elsevierdigitaledition.com/main/119/index…

Thrilled to share our latest research, SIMPLE-seq, a high-throughput single cell method enabling simultaneous profiling of 5mC and 5hmC. Our SIMPLE-seq work is online now @NatureBiotech! Huge thanks to the exceptional mentor @YiChengqi @chenxuzhu, and all lab members!
Simultaneous single-cell analysis of 5mC and 5hmC with SIMPLE-seq - @PKU1898 @chenxuzhu @YiChengqi go.nature.com/49NeNFN
We are excited to start the project to identify the molecular changes of aging with single-cell multiomics! If you are interested in tech dev in scMultiomics, epigenomics, DNA damages & aging, please join us: jobs.silkroad.com/NYGenome/Caree…
Chenxu Zhu, PhD, of @nygenome and @WeillCornell, has received the prestigious @NIH Director's New Innovator Award from the @NIH_CommonFund. Congratulations to Dr. Zhu on this well-deserved accomplishment! #NIHHighRisk bit.ly/3S18hWj
Weill Cornell Medicine's Dr. Chenxu Zhu, also of @nygenome, has received the @NIH Director's New Innovator Award from @NIH_CommonFund to develop tools to study gene expression and changes in brain cells of preclinical models during aging. #NIHHighRisk bit.ly/48FhMQP
We are super excited to receive the DP2 award. Thank you, #NIHHighRisk and @NIGMS, for supporting our vision of studying the biology of brain aging with innovative single-cell multi-omics technologies.
Check out the tools being developed to better understand #epigentics and #geneexpression as it relates to aging by #NIHHighRisk New Innovator Awardee Dr. Zhu of @nygenome and @WeillCornell
🍺I am so excited that my first paper is online (nature.com/articles/s4159…)! Joint profiling of histone modifications and transcriptome are critical to understand the active or repressive gene regulatory network. We therefore develop Droplet Paired-Tag,
Dr. Chenxu Zhu's research group at the New York Genome Center (NYGC) and Weill Cornell Medicine (WCM) is looking for motivated postdoctoral researchers interested in single-cell genomics and gene regulation. jobs.silkroad.com/NYGenome/Caree…
Honored to participate in this symposium and receive Travel Awards! Thank you, Prof. Li, for kind invitation and organization!
Thank you University of Rochester and Zhejiang University (ZJU) for making our Young Investigator Symposium on RNA Biology a success with 24 speakers & 173 attendees. Congrats to Li Yanqiang, Song Jinghui, and Fang Junnan on winning Travel Awards to visit ZJU RNA Center at Yiwu!
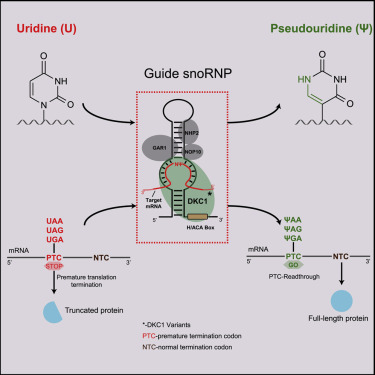
Online Now: CRISPR-free, programmable RNA pseudouridylation to suppress premature termination codons dlvr.it/SfMKvd
I am thrilled to announce that I will join @nygenome and @WeillCornell in 2022! I cannot wait to join these fantastic communities and work with the amazing scientists!! My future lab will focus on single-cell tech dev, and postdoc positions are available: nygenome-openhire.silkroad.com/epostings/inde…