Genome in a Bottle
@GenomeInABottle
The Genome in a Bottle Consortium develops reference materials, reference data, and reference methods needed to benchmark human genome sequencing
Our v4.2.1 small variant benchmarks using long and linked reads for 7 @GenomeInABottle samples, and their use in the @precisionfda Truth Challenge, now published in @CellGenomics. Thanks to the many contributors to these 2 papers! doi.org/10.1016/j.xgen… doi.org/10.1016/j.xgen…
Nice to see the impact of this important work. This benchmark set (v4.2.1) drove a huge amount of improvement sequencing. All seq instruments use it to quantify performance. Best credit is to @GenomeInABottle, who drove the field forward both in this work and over a decade.
Benchmarking challenging small variants with linked and long reads by @acarroll_ATG et al. (62 citations) hubs.li/Q035ZMbM0 Top-cited genomics research published in @CellGenomics
Our first curated draft somatic structural variant benchmark for the new GIAB PDAC tumor cell line HG008-T is at ftp-trace.ncbi.nlm.nih.gov/ReferenceSampl…, based on extensive short+long read sequencing data described in doi.org/10.1101/2024.0…. Feedback to improve future versions is very welcome!
We are pleased to announce the release of the Genome in a Bottle (@GenomeInABottle) Problematic Regions tracks for the hg38 and hs1 human assemblies. Learn more about the release from the following news post: genome.ucsc.edu/goldenPath/new…
don't use hg38.fa as-is. checkout the references 😜 here: ftp-trace.ncbi.nlm.nih.gov/ReferenceSampl… rendered the ipynb (not mine) here: gist.github.com/brentp/1935e9b… in short, use: GRCh38_GIABv3_no_alt_analysis_set_maskedGRC_decoys_MAP2K3_KMT2C_KCNJ18.fasta.gz other updates on the best hg38 reference?
We realized they could be mosaic variant positions. Fortunately, NIST @GenomeInABottle released a set of annotated mosaics in the cell lines we were looking at. Plotting those examples revealed they overlap the cluster we inspected.
The @GenomeInABottle genomic stratifications resource is published in @NatureComms: nature.com/articles/s4146… Stratifications reveal key insights into precision and recall of variant calling across different genomic contexts! Great team work by Nate Dwarshuis, Justin Zook & others
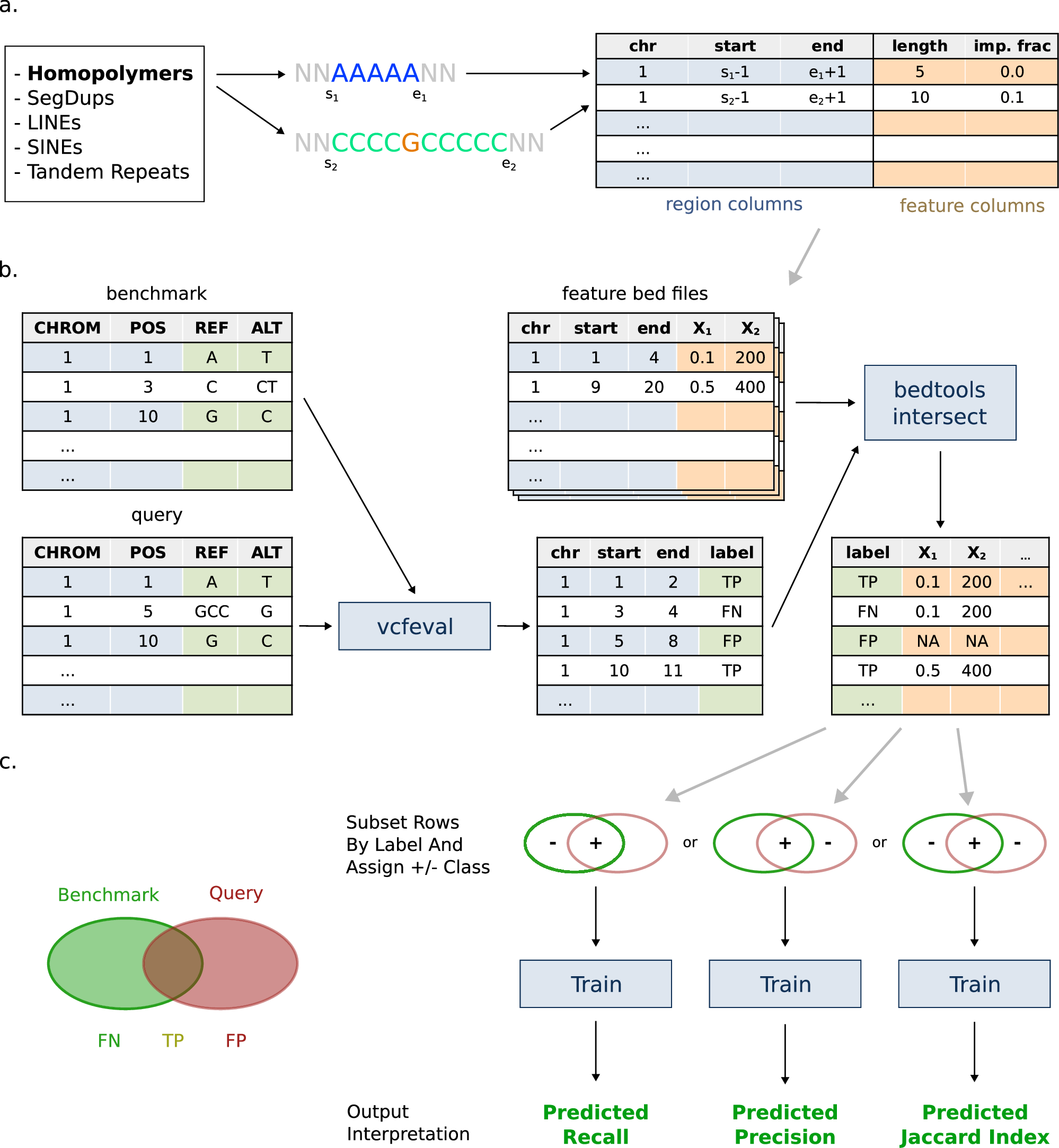
An interesting new method to use interpretable machine learning with GIAB benchmarks to understand how variant calling errors are correlated with different types of genomic repeats nature.com/articles/s4200…
Interested in benchmarking computational methods in computational biology, generally? Save the date, submit an abstract, join us next March in Ascona! Conference website: sites.google.com/view/ascona2025 Please retweet and/or tell your colleagues!
Extensive WGS for 1st @NIST GIAB tumor cell line, consented for public genomic data doi.org/10.1101/2024.0…. PDAC tumor/normal pair with data from @nanopore @PacBio's HiFI&Onso @illumina @ElemBio @UltimaGenomics @bioskryb @ArimaGenomics @PhaseGenomics @bionano @KromatidInc
📢 By popular demand, you can now add tracks to IGV Desktop directly from 42basepairs!
Tandem repeat @GenomeInABottle benchmark @NatureBiotech out today: rdcu.be/dFQNN . Characterization of 1.7 million TR + benchmark variants + new method to overcome var. representation issues! Great work lead by Adam E. @BCM_HGSC & great collab. from so many! (1/4)
Want to learn more about the tricks and tips for generating ONT Ultra-long DNA? The T2T Consortium and UCSC SeqTech Center are hosting a Technology-focused Webinar next Thursday with a group of experts. Register today: bit.ly/3PFTa2f @aphillippy
SplitThreader, my cancer SV viz tool, is 8 years old. I have some ideas for giving it a makeover that would make it much more lightweight and easy to prepare input files for. Please reply if you are interested so I can tell if I should prioritize this! splitthreader.com/vis.php?code=e…
Our 10th LR seminar @BCM_HGSC at 22nd March 10am CT! Come join us and listen to new advancements on LR from @mitenjain @tycheleturner @GenomeInABottle @VanessaPorter Register: hgsc.bcm.edu/LongReadSeminar @bcmhouston @bcmgenetics @PacBio @nanopore @RiceCompSci @TXMedCenter
Two other resources of note for IGV: - Interactive guide to getting started with IGV: sandbox.bio/tutorials/igv-… - The supplement to this article, which is an incredibly comprehensive visual guide to reviewing somatic variants: ncbi.nlm.nih.gov/pmc/articles/P…
A key skill in bioinfo is assessment of raw seq data. Tools like IGV are useful to view aligned reads & variants. I've manually reviewed 1000s of variants and it's worth knowing what a real variant looks like! From Sirisha in the @nanopore #EPI2ME team: labs.epi2me.io/reviewing-bam
Preprint on Exploring gene content with pangenome gene graphs: arxiv.org/abs/2402.16185. It describes pangene for building gene graphs and for calling gene-level variations which can be found at pangene.bioinweb.org. Pleasant collaboration with @maxgmarin and @MahaFarhat.
Thank you Director Green! BioDIGS has truly been a highlight of my career. It starts with collecting soil from all over the United States and then diving deep into the informatics to study the genomic diversity of the world around us. schatz-lab.org/presentations/…
A highlight of the morning session at #AGBT2024 was @mike_schatz’s talk, in which he described a remarkably creative new program (BioDIG) that involves getting students involved in microbiome research of soil. Kudos to Michael for his outstanding leadership of this effort!
New vcfdist paper on bioRxiv! Key Takeaways: 1) Jointly evaluating small and structural variants decreases measured FN+FP by 20-50% 2) 43-92% of phasing flip "errors" are FP due to variant representations More below Code: github.com/TimD1/vcfdist Paper: biorxiv.org/content/10.110…