UCSC Genome Browser
@GenomeBrowser
The UCSC Genome Browser is a public, freely available, open-source web-based graphical viewer for the display of genome sequences and their annotations.
We are happy to announce the release of the Bionano DLE-1 track for human assemblies hg38 and hg19. These tracks show the CTTAAG sites used by the Bionano Optical Genome Mapping system, an assay to detect structural variants. Learn more at: genome.ucsc.edu/goldenPath/new…
New ENCODE4 long-read RNA-seq transcripts track for hg38 and mm10. Triplets (e.g. [1,1,3]) indicate start site, exon combination, and stop site for each transcript. Enrichment scores show how these change across tissue and cell line samples. Read more: genome.ucsc.edu/goldenPath/new…
![GenomeBrowser's tweet image. New ENCODE4 long-read RNA-seq transcripts track for hg38 and mm10. Triplets (e.g. [1,1,3]) indicate start site, exon combination, and stop site for each transcript. Enrichment scores show how these change across tissue and cell line samples.
Read more: genome.ucsc.edu/goldenPath/new…](https://pbs.twimg.com/media/Gv_8O-kXcAADW7n.png)
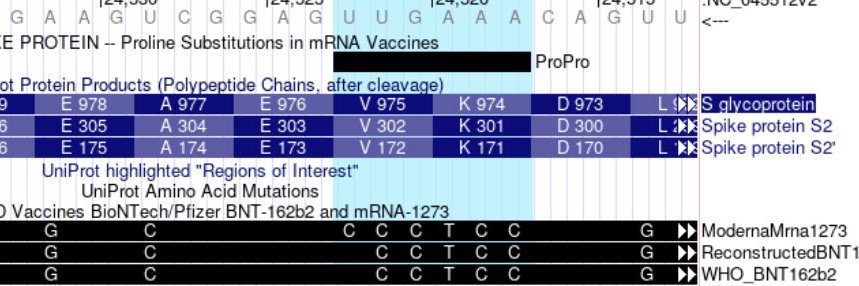
We have two new pathogenicity prediction score tracks available in our Deleteriousness Predictions super track: MCAP and MutScore! Both are aimed at interpreting the pathogenicity of variants in a clinical setting. See our news for more: bit.ly/UCSCmutScoreMC…

We are happy to announce the release of the Unusually Conserved Regions track for GRCh38/hg38. For more information about the release, please read the news announcement: genome.ucsc.edu/goldenPath/new…
We turn 25 today! July 7th marks the 25th anniversary of the human genome going online and the start of the UCSC Genome Browser. Then vs. now, we have 165k monthly visitors, and our codebase is over three million lines of code. See our news for more: bit.ly/genomeBrowser2…

We have a new training page with updated tutorials covering our most popular tools. Each includes an annotated screenshot, guided walkthroughs, and an interactive click-through tutorial. See our news for more: bit.ly/genomeBrowserT…

We are pleased to announce the release of the All GENCODE tracks for hg19, hg38, and mm39. These tracks correspond with the @ensembl 114 release. Learn more about the release from the following news post: genome.ucsc.edu/goldenPath/new…
We are happy to announce the release of the EVA SNP Release 7 tracks, now available for 40 assemblies and covering nearly 910 million variants. Learn more at genome.ucsc.edu/goldenPath/new….

Are you at #eshg2025? Visit us at booth 226! We will also be co-presenting a workshop on Monday, 5/26, with @ensembl at 14:15.


We are excited to announce the release of the Varaico tracks for human assemblies hg38 and hg19. Varaico stands for "Variation Research Advancing Insight in Complex Organisms". Varaico was created using literature mining, similar to AVADA. Learn more at: genome.ucsc.edu/goldenPath/new…

New tracks now available: GENCODE "knownGene" V48 for human (hg38/hg19) and VM37 for mouse (mm39). Read more about this release in this news post: genome.ucsc.edu/goldenPath/new…
We are happy to announce an update to the VISTA Enhancers tracks for hg38, hg19, mm39, and mm10. Learn more about the release from the following news post: genome.ucsc.edu/goldenPath/new…
Announcing the new Pseudogenes track for hg38! The composite track contains pseudogene predictions and their corresponding parent genes, as identified by PseudoPipe. This release includes the Pseudogene Parents and Pseudogenes tracks. Learn more at genome.ucsc.edu/goldenPath/new….

A new hub has been added to the Public Hubs listing, EPIration - an atlas of candidate enhancer-gene interactions in human tissues and cell lines. View this hub using the following URL: genome.ucsc.edu/cgi-bin/hgHubC… We want to thank the @GuigoLab for making this data available.
We are excited to announce the release of two denovo-db tracks for hg19/GRCh37. denovo-db is a curated database of germline de novo variants in the human genome, defined as variants present in children but absent in their parents. Learn more at: genome.ucsc.edu/goldenPath/new…
We are excited to share AlphaMissense tracks for the hg38/hg19 human assemblies. AlphaMissense scores, generated by @GoogleDeepMind, predict the pathogenicity of missense variants for all possible single AA substitutions in the human proteome. Learn more: genome.ucsc.edu/goldenPath/new…

We are pleased to announce the release of the MITOMAP track for hg38/hg19. MITOMAP is a human mitochondrial genome database. This release includes the MITOMAP Control and Coding Variants track & the MITOMAP Disease Mutations track. Learn more at genome.ucsc.edu/goldenPath/new….
