
Matthew Parker
@bioinfomatt
HCPC Registered Clinical Bioinformatician. Director, Clinical Bioinformatics Software @nanopore. Views are my own.
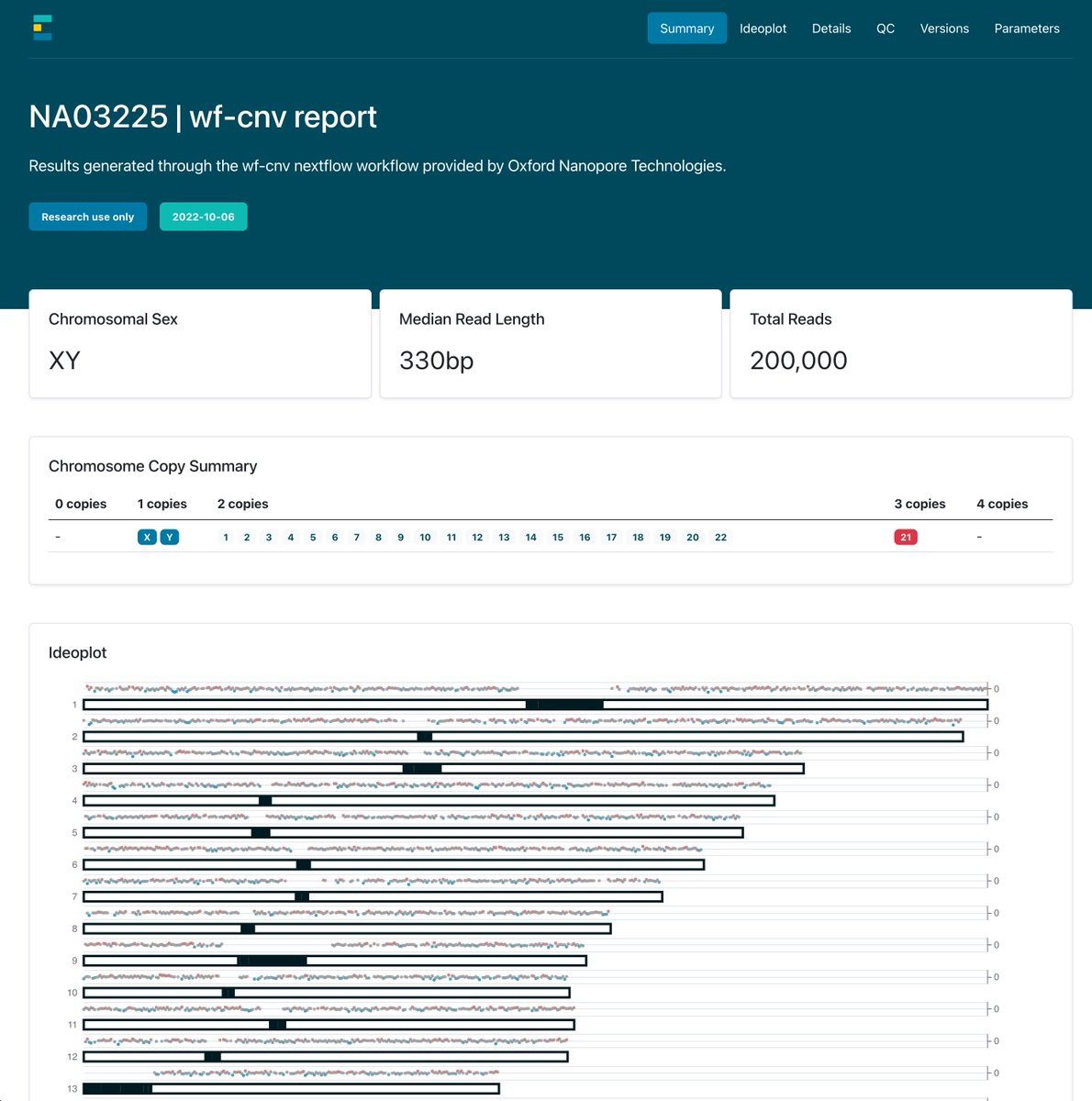
We've released a new workflow that allows the calling of copy number variation from @nanopore human sequencing data. This uses QDNASeq & settings honed by the applications team to produce the best results. Also uses our new report framework. Check it out! labs.epi2me.io/copy-number-ca…

EPI2ME CLOUD IS LIVE our overhauled @nanopore EPI2ME Desktop crams in delightful UX which seamlessly supports our entire suite of bioinformatics workflows on your computer, or in a private stack on our new cloud infrastructure GIVE IT A SPIN labs.epi2me.io/downloads/
We continue to commit to a unified EPI2ME experience. We've launched the #EPI2ME cloud enabling EPI2ME in the cloud or locally - data analysis for all. #nanoporeconf
🚀 Check out our latest blog post by Chris Wright and @PaoloDiTommaso exploring EPI2ME Workflows — a collection of professionally maintained, high-quality @nextflowio pipelines from the team at @nanopore 👉 Read now: hubs.la/Q02TWSD10 #Nextflow #EPI2ME #Seqera
It’s time for the most energetic of #bioinformatics updates with our very own @SAGRudd ! #EPI2ME #nanopore
We continue to commit to a unified EPI2ME experience. We've launched the #EPI2ME cloud enabling EPI2ME in the cloud or locally - data analysis for all. #nanoporeconf
BugSeq is delighted to announce that we are now listed on the @nanopore compatible products page. This listing provides users with enhanced confidence in the performance of BugSeq’s analysis platform with ONT sequencing workflows.
It's Oxford @nanopore's first #BiopharmaDay at the MANIFICIENT @BPLBoston! A full house & full day of fantastic talks by representatives from #biopharma discussing how they are using, or planning to use, the power #nanopore #directRNA & DNA sequencing in R&D and #QC #nanoporeconf
Masterclass demos by @bioinfomatt and Tim Walker on the Q-line plasmid and direct mRNA assays - killer apps for biopharma QC!
Was fortunate enough to test this early on and it will really be a game changer especially for using @nanopore sequencing with high school classes. Genome assembly, metagenomics analysis etc made easy with no requirement for HPC or complex software installations #nanoporeconf
We continue to commit to a unified EPI2ME experience. We've launched the #EPI2ME cloud enabling EPI2ME in the cloud or locally - data analysis for all. #nanoporeconf
We all know @philres1 is awesome, but man he smashed this talk. Complex @nanopore multiomic data made so easy and impactful. #NanoporeConf
Awesome talk at the @nanopore conference about multiomics unravelling of the Rett syndrome
Dorado v0.8.0 #ncm24 release! - v5.0 DNA transformer model performance improved for A100/H100 via kernel fusing, quantisation and leveraging i8 tensor cores. - v5.1 RNA models with updated mod calling for m6A, pseU, and brand new calling of Inosine and m5C. - Improved…
We couldn’t be more excited to get this out of the door to y’all!
live from #NCMBOS2024 EPI2ME Cloud is finally released for everyone! we can’t wait to see what incredible bioinformatics analysis you all will do on the platform 🤘🚀
We continue to commit to a unified EPI2ME experience. We've launched the #EPI2ME cloud enabling EPI2ME in the cloud or locally - data analysis for all. #nanoporeconf
We continue to commit to a unified EPI2ME experience. We've launched the #EPI2ME cloud enabling EPI2ME in the cloud or locally - data analysis for all. #nanoporeconf
Have the Epi2Me software for analysis. Very helpful
Super exciting to see what the future holds for nanopore beyond DNA and RNA sequencing nature.com/articles/s4158…
In this paper we report on 20 retrospective and 50 prospective intraoperative patient samples, providing a classification within 2 hours of sample receipt and a full molecular classification within 24 hours using the unique real time features of @nanopore sequencing.
Adding another to the collection! @nanopore #NCM2024 #nanoporeconf #EPI2ME

Excited to receive our @nanopore Mk1D today! Looking forward to getting it up and running next week and then getting it out on fieldwork soon!
Introducing ROBIN. Rapid nanopOre Brain intraoperatIve classificatioN. ROBIN builds on work by many groups and shows how we could reduce turnaround time for full molecular classification of CNS tumours in NHS clinical settings from weeks to hours. medrxiv.org/content/10.110…

