
Sina Majidian
@SinaMajidian
On the academic job market| http://bsky.app/profile/sinamajidian.bsky.social | Comparative pangenomics at scale | PostDoc at Langmead Lab | Prev. UNIL/SIB/WUR
FastOMA is out now in Nature Methods 🎉: nature.com/articles/s4159… A new orthology inference algorithm that scales linearly and is highly accurate. FastOMA can process all >2000 eukaryotic UniProt ref proteomes <24 hours 🚀. Try it out at github.com/DessimozLab/fa…

Proud moment :D. I think this is the first study that uses both TRUST4 (for TCR assembly) and T1K (for HLA genotyping).
T cell receptor sharing in hypersensitivity pneumonitis biorxiv.org/cgi/content/sh… #biorxiv_immuno
📖 Read more on how these advancements are reshaping genomics research: annualreviews.org/content/journa…
Anyone know what's going on with AcademicJobsOnline (@ajobsonline)? Their site has been down for more than a week.
Exciting News! 🚀 We want to hear from YOU! 📢 Take a moment to share your experience in biomedical data analysis and help shape the future of AnVIL. Complete the State of the AnVIL Community Poll here and enter to win a gift card: bit.ly/sota2025
How are different #orthology methods related and how well do they perform? Read about this and a new orthology benchmark in "New developments for the Quest for Orthologs benchmark service" academic.oup.com/nargab/article…
Have you checked out our behind-the-paper blog post on FastOMA? Read it here go.nature.com/4a3V9q3 I’ll give you a quick intro to orthology, a bit of history of orthology methods, and the story of how FastOMA came to life—and how it works!
From @SinaMajidian, @DessimozLab and colleagues, FastOMA achieves fast and accurate orthology inference, with linear scalability. nature.com/articles/s4159…
Interesting talk by Benjamin Parks at #biodata24, showing how creative ideas in data compression can improve performance of single cell analysis github.com/bo1929/krepp deployed in Seurat satijalab.org/seurat/article… achieving upto 60x speedups compared to ArchR/SnapATAC2 #cshldata24

Check out our new work led by @ashton_omdahl discussing matrix factorization across GWAS, accounting for sample overlap. Factors with varying polygenicity, enrichment for cell type / developmental stage... Ashton is speaking at #biodata24 today! biorxiv.org/content/10.110…
Excited to share our work on splice site prediction! Stop by poster #66 tonight to learn more about Splam and OpenSpliceAI. 🔗Read the paper here: doi.org/10.1186/s13059…. #biodata24
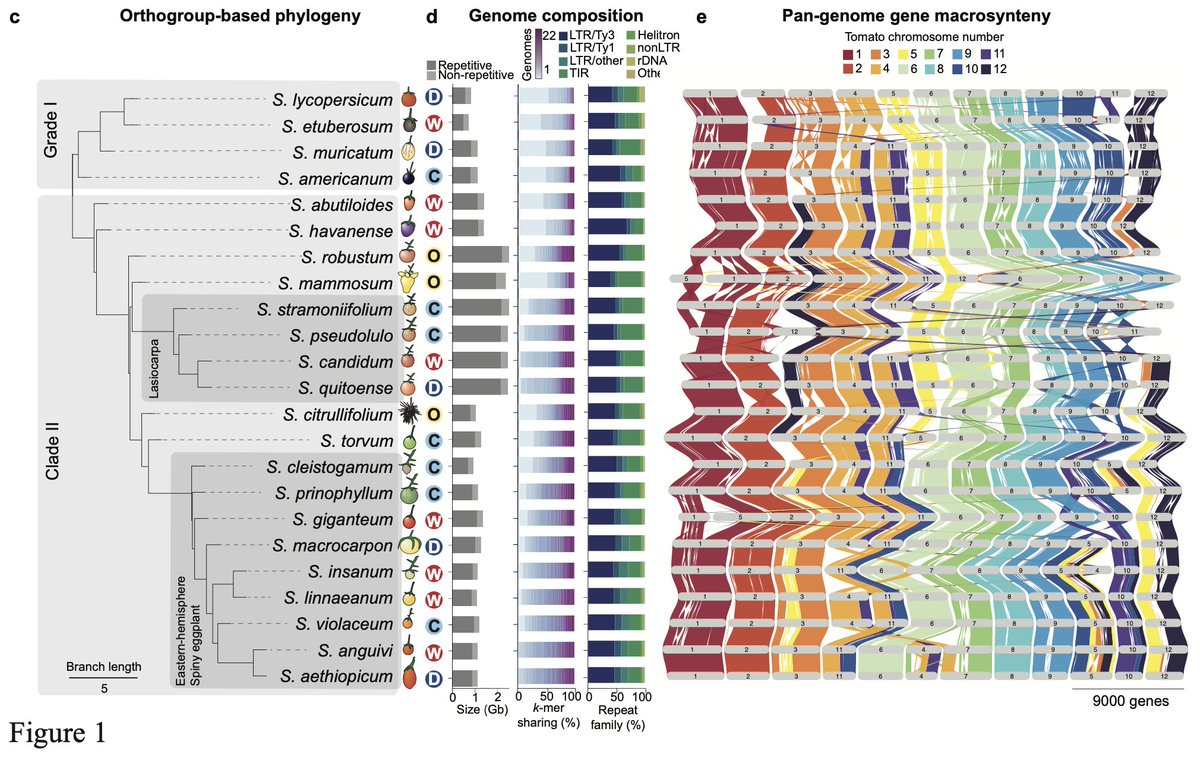
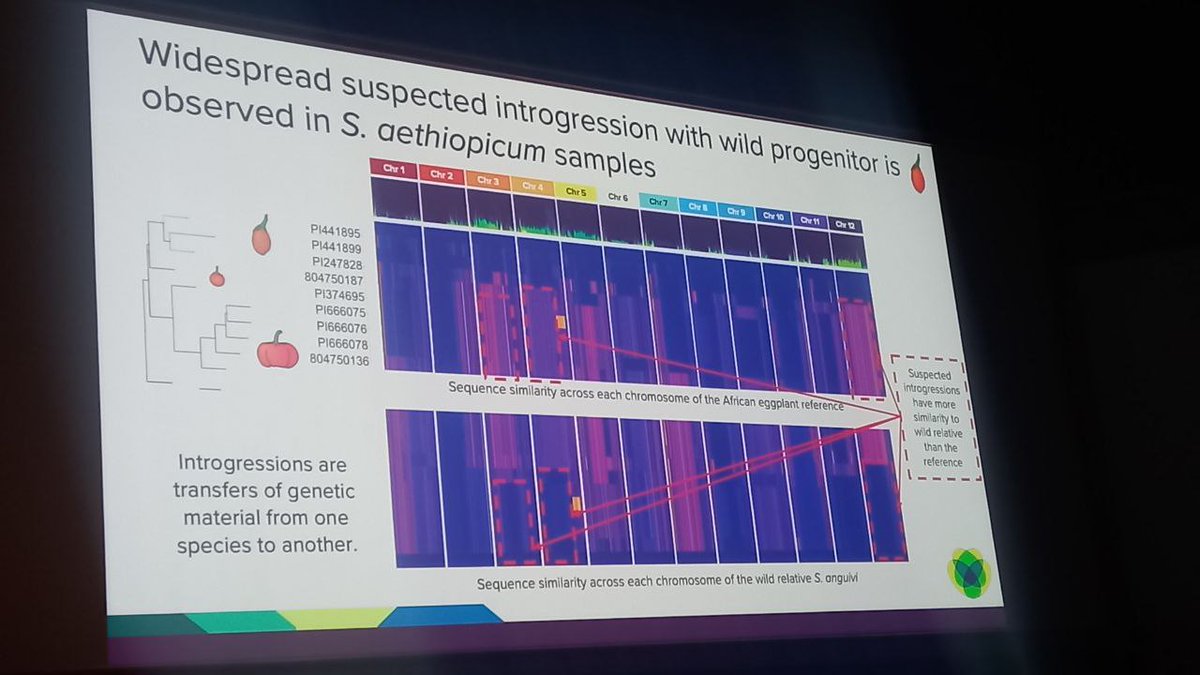
Great talk by Katharine Jenike at #biodata24 on investigating paralog evolution in tomato and eggplant using a pangenomic approach, revealing widespread introgressions biorxiv.org/content/10.110… Amazing synteny viz with github.com/jtlovell/GENES… #cshldata24
Looking for a postdoc in machine learning, data science, AI? We are launching a postdoc program at JHU! Candidates in foundations/methods or applications welcome! Please share/RT! (For my friends & followers, yes this includes genomics and health applications)
We’re thrilled to announce the #HopkinsDSAI Postdoctoral Fellowship Program! We’re looking for candidates across all areas of data science and AI, including science, health, medicine, the humanities, engineering, policy, and ethics. Apply today! ai.jhu.edu/postdoctoral-f…
Cool idea of universal single-isoform genes (BUGSI) to assess RNA data as part of the SQANTI ecosystem. A great talk by @anaconesa @ConesaLab at #cshldata24 SQANTI-Sim: link.springer.com/article/10.118… #biodata24 SQANTI3 for quality control long-read transcriptome github.com/ConesaLab/SQAN…
Great talk by @cumbofabio from @ClevelandClinic at #biodata24 on MetaSBT for characterizing microbial genomes via hierarchical structure of sequence bloom trees. The ability to update the index and adding new genomes is amazing! github.com/cumbof/MetaSBT… #cshldata24
Biological Data Science starts tonight! Use #cshldata24 this week for all of your posts 🧬
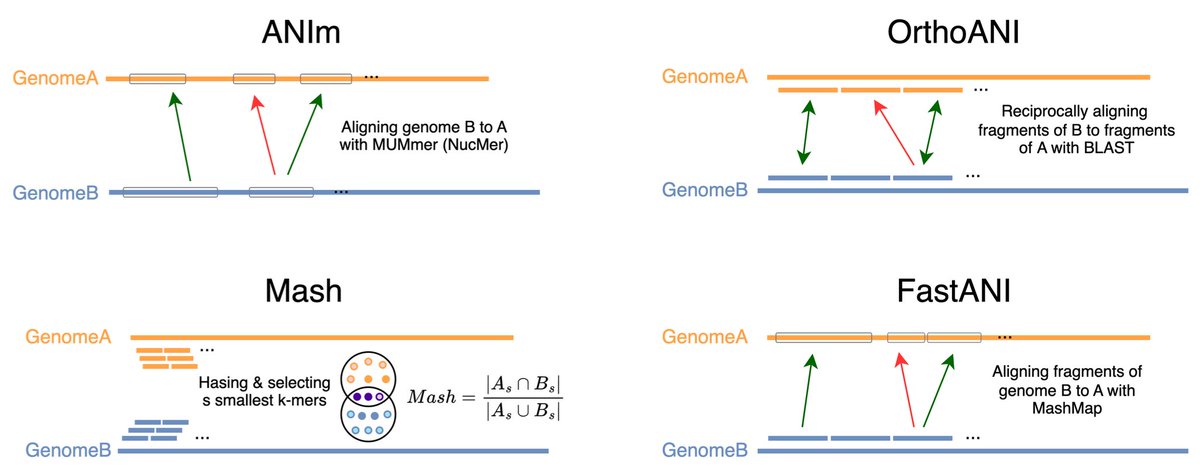
Heading to Cold Spring Harbor to present a poster on benchmarking average nucleotide identity! "ANI, are you OK? Challenges for k-mer-based estimation of evolutionary distances" at Biological Data Science #cshldata24 !
Learn more about the stratifications used here and more in our recent paper at nature.com/articles/s4146…