Ben Kleinstiver
@BKleinstiver
Associate Professor @CGM_MGH at @MGH_RI & @HarvardMed Genome editing / Protein eng. / Molecular medicine 🇨🇦🧬 Kayden-Lambert MGH Research Scholar '23-28
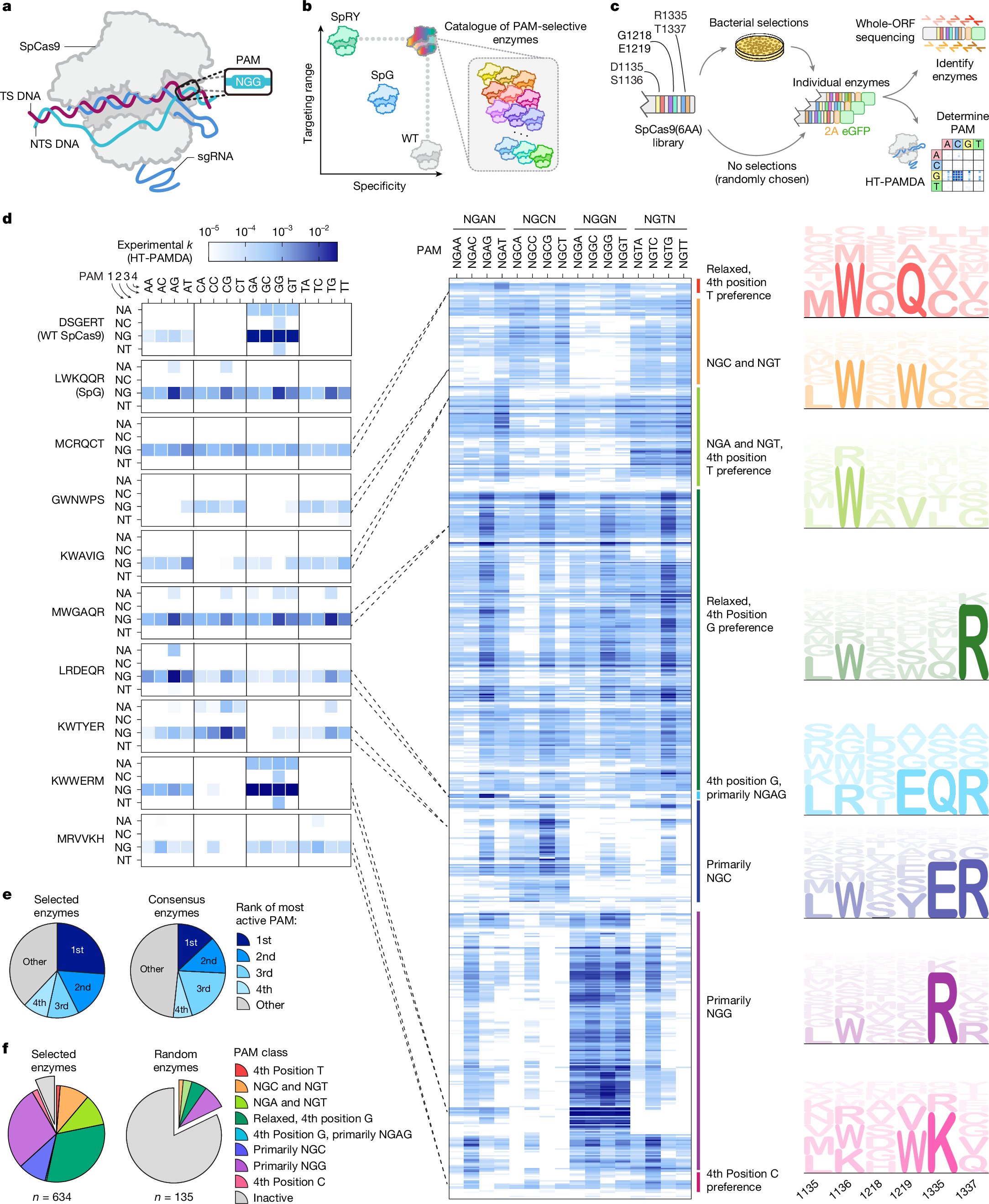
In @nature we describe the use of scalable #proteinengineering & #machinelearning to predict millions of bespoke CRISPR-Cas9 enzymes, offering safer & more genome editing tools. 🧬🖥️ @RachelSilvers11 @CGM_MGH @MGH_RI @MassGeneralNews @harvardmed nature.com/articles/s4158…
1/16 New pre-print from the Sternberg Lab! We uncover how temperate phages can use RNA-guided transcription factors to remodel the flagellar composition of their bacterial host and enhance their fitness. Find the preprint and full story here: tinyurl.com/mshwjd77
We’re @BlaineyLab sharing a major update to CROPseq-multi, our versatile system for CRISPR screens that is compatible with individual and combinatorial perturbations, diverse SpCas9-based technologies, and multiple high-content, single-cell readouts. biorxiv.org/content/10.110…
A triumph of science, a miracle of medicine - built on decades of public investment into the US basic and translational biomedical research ecosystem. Magnificent effort led by Drs Rebecca Ahrens-nicklas (CHOP) and Kiran Musunuru (Penn) + IGI + Danaher. innovativegenomics.org/news/first-pat…
"Does it work in human cells?" is a question I've gotten a lot this past year. Excited to share our latest work on bridge recombinases! It's been a lot of fun to lead this team effort. If you are excited to hear more, join for my thesis defense tomorrow at 3pm PT. Links below:
Genomes encode biological complexity, which is determined by combinations of DNA mutations across millions of bases In new @arcinstitute work, we report the discovery and engineering of the first programmable DNA recombinases capable of megabase-scale human genome rearrangement
1/10 Today in @ScienceMagazine in collaboration with @liugroup we report the development of a laboratory-evolved CRISPR-associated transposase (evoCAST) that supports therapeutically relevant levels of RNA-programmable gene insertion in human cells. drive.google.com/file/d/1I-UbCR…
Breaking News: A baby with a rare disorder made medical history by receiving the first custom gene-editing treatment. The technique used has the potential to help people with thousands of other uncommon genetic diseases. nyti.ms/4j3xGs8
Today in @ScienceMagazine we report the development of a laboratory-evolved CRISPR-associated transposase (evoCAST) that supports therapeutically relevant levels of RNA-programmable gene insertion in human cells, a collaboration with @SternbergLab. 1/13 drive.google.com/file/d/1I-UbCR…
BREAKING: Last August, scientists secretly began creating a custom #CRISPR therapy to fix a mutation responsible for one baby’s debilitating disease. He was treated in February and doing better. My deep dive in @endpts has the details. endpts.com/philadelphia-d…
Congratulations to @davidrliu on being named to the #TIME100Health list — honoring the 100 most influential people in health today. A well-deserved recognition!
TIME's new cover: Meet this year's #TIME100Health, a community of leaders—scientists, doctors, advocates, educators, and policy-makers, among others—who are changing the health of the world. ti.me/43802LZ
We’re excited to share new work on the continuous in vivo evolution of gene libraries towards arbitrary functions. Led by superstar Olek Pisera, our experiments show that gene evolution is highly versatile...biorxiv.org/content/10.110…
Machine Learning Engineers Bespoke Cas9 Enzymes for Gene Editing Read more of what @BKleinstiver and @RachelSilvers11 from HMS had to say in an interview with GEN's @xiaofei_lin. @MassGeneralNews @harvardmed hubs.li/Q03jt42N0
Excited to share my PhD work… and more CRISPR-Cas9 PAM variants than anybody asked for
In @nature we describe the use of scalable #proteinengineering & #machinelearning to predict millions of bespoke CRISPR-Cas9 enzymes, offering safer & more genome editing tools. 🧬🖥️ @RachelSilvers11 @CGM_MGH @MGH_RI @MassGeneralNews @harvardmed nature.com/articles/s4158…