
Taekjip Ha
@taekjip
Biophysicist. Howard Hughes Medical Institute. Harvard Medical School. Boston Children’s Hospital. Father of three dragons. Occasional manuscript reviewer.
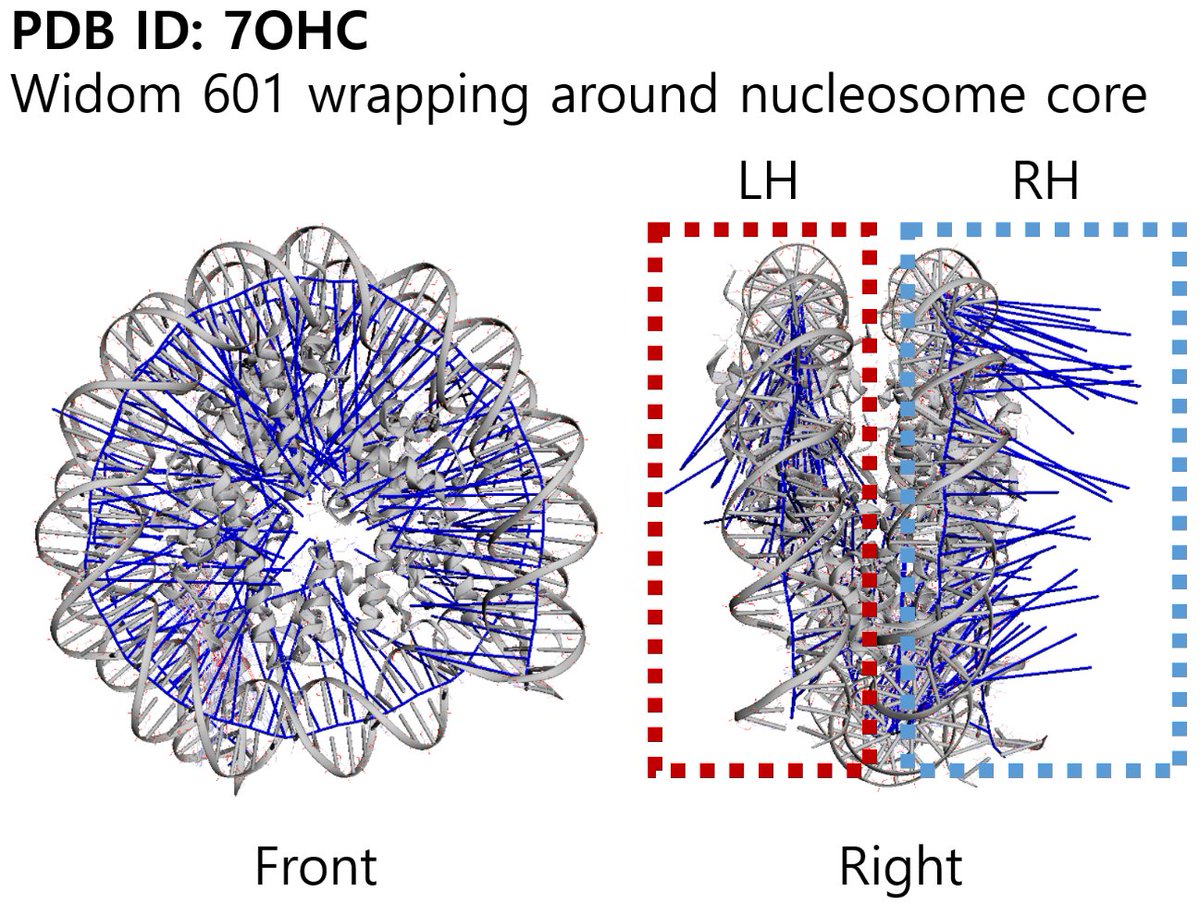
We have a web interface for predicting DNA cyclizability. Entering a PDB ID shows the preferred bending direction around a protein. Widom 601 DNA has strong and weak sides, and indeed intrinsic bending is toward the nucleosome center for the strong side (LH) only. Plz try it out!
Johns Hopkins neuroscientist Uli Mueller came to the U.S. for the chance to do research he couldn't do anywhere else. Now, he’s concerned those opportunities are slipping away as funding becomes less reliable. Read more: hub.jhu.edu/2025/05/21/us-… #ResearchSavesLives
A conserved coupling of transcriptional ON and OFF periods underlies bursting dynamics | Nature Structural & Molecular Biology nature.com/articles/s4159…

So proud of the smashing success of the International Course in Biophysics co-organized with my @berkeleyMCB colleague Carlos Bustamante @BustamanteLab and University of Salamanca @usal biophysal.org Course ends with a Keynote from Nobel Laureate Richard Henderson🥇
🎉 Kicking off #Nucleic25 with an inspiring talk from @taekjip on homology search in 3D genome - setting the tone for an exciting few days ahead! 🧬 🎉Our amazing poster presenters took the stage to introduce themselves and their work 🎉We wrapped up the day with a dinner full…
Thanks TJ @taekjip for giving me the opportunity to work on this, “Mechanistic insights into direct DNA and RNA strand transfer and dynamic protein exchange of SSB and RPA”academic.oup.com/nar/article/53…
high-throughput single-molecule platform to study DNA supercoiling effect on protein–DNA interactions academic.oup.com/nar/article/53…

DNA origami-based platform for multi-axial single-molecule force spectroscopy reveals hidden dynamics in Holliday junctions biorxiv.org/content/10.110…
Non-disruptive 3D profiling of combinations of epigenetic marks in single cells biorxiv.org/content/10.110…
Congrats to HHMI Freeman Hrabowski Scholars Chantell Evans, Josefina del Mármol, Yvette Fisher and Theanne Griffith—named 2025 McKnight Scholars for their demonstrated commitment to neuroscience and dedication to mentoring others! 🌟 Congratulations to all the awardees! Read…
Bringing the Genetically Minimal Cell to Life on a Computer in 4D biorxiv.org/content/10.110…
The secret to how nucleosomes shape the genome | Journal Club | PNAS pnas.org/post/journal-c…

Proud to announce our new technique for tracking relative B cell clonality in situ with B Cell Receptor MERFISH (BCR-MERFISH)! Congratulations to Evan Yang and our colleagues in the Carroll laboratory! Check out our bioRxiv: biorxiv.org/content/10.110… 1/9
The Theoretical and Computational Biophysics Group is pleased to announce the 63rd "Hands-on" Workshop on Computational Biophysics (tcbg.illinois.edu/Training/Works…) to be held August 4-8, 2025 at Auburn University in Auburn, Alabama.
Commemorate the life and science of our dear departed colleague Jon Widom - this Thurs May 15 1230 pm in Pancoe Auditorium @NUmolbiosci @NorthwesternU - our distinguished Widom Lecturer is Prof. Lucy Bai @penn_state - Chromatin Accessibility and the 3D Genome
New preprint on single-molecule mechanostructural fingerprinting! doi.org/10.1101/2025.0… We use DNA nanoswitch calipers to characterize the geometry and mechanical stability of telomeric DNA G-quadruplexes. #Biophysics #SingleMolecule #Mechanobiology #DNAnanotech
Amazing work from the HaLab, and this interplay between biophysical properties and genome organization are so cool! Congrats to Sangwoo and an amazing team.
Check out this tour de force study by Sangwoo Park and his incredible team of collaborators. Biophysical properties of single nucleosomes are responsible for A and B compartments in cell nucleus.
Check out this tour de force study by Sangwoo Park and his incredible team of collaborators. Biophysical properties of single nucleosomes are responsible for A and B compartments in cell nucleus.
"Native nucleosomes know where to go!” Our Condense-seq paper is now published in Nature! 📄 nature.com/articles/s4158… I’m deeply grateful to my PhD advisor @taekjip and our incredible collaborators @MuirLab , @binzmit , Erika Pearce, and Ben Garcia lab!!
"Native nucleosomes know where to go!” Our Condense-seq paper is now published in Nature! 📄 nature.com/articles/s4158… I’m deeply grateful to my PhD advisor @taekjip and our incredible collaborators @MuirLab , @binzmit , Erika Pearce, and Ben Garcia lab!!
How do nucleosomes encode genome organization principles?@Nature @HopkinsMedicine "Native nucleosomes intrinsically encode genome organization principles" The research investigates whether individual nucleosomes contain sufficient intrinsic information for 3D genomic…
