Karthik Guruvayurappan
@somet3000
First-year PhD Student at Tri-I Computational Biology and Medicine program (Weill Cornell, Memorial Sloan Kettering Cancer Center, Rockefeller University)
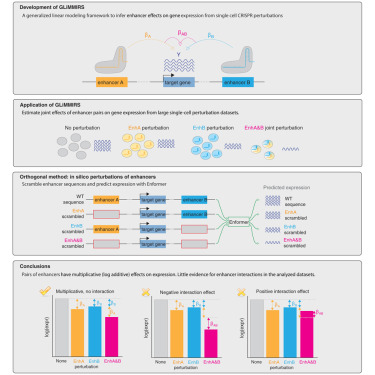
Happy to share that the paper on enhancer interactions led by @zrcjessica and I from my time in the @GrahamMcVicker lab is now published in Cell Genomics! cell.com/cell-genomics/… A thread on some of the new analyses below: (1/n)
Our work on "Evaluating the representational power of pre-trained DNA language models for regulatory genomics" led by @AmberZqt with help from @NiraliSomia & @stevenyuyy is finally published in Genome Biology! Check it out! genomebiology.biomedcentral.com/articles/10.11…
Do current genomic language models (pre-trained on whole genomes) learn a foundational understanding of biology in the non-coding region of human genomes? A new evaluation led by @AmberZqt suggests not yet! 1/N paper: biorxiv.org/content/10.110…
I am excited to introduce mRNABench, a comprehensive benchmarking suite that we used to evaluate the representational capabilities of 18 families of nucleotide foundation models on mature mRNA specific tasks. Paper: doi.org/10.1101/2025.0… Code: github.com/morrislab/mRNA… A 🧵
We're excited to release 𝐦𝐑𝐍𝐀𝐁𝐞𝐧𝐜𝐡, a new benchmark suite for mRNA biology containing 10 diverse datasets with 59 prediction tasks, evaluating 18 foundation model families. Paper: biorxiv.org/content/10.110… GitHub: github.com/morrislab/mRNA… Blog: blank.bio/post/mrnabench
🧬 Excited to open-source Biomni! With just a few lines of code, you can now automate biomedical research with AI agent! We are releasing Biomni A1 (agent) + E1 (env) with 150 specialized tools, 59 databases, and 105 software. E1 is our first attempt at curating the bio-agent…
want to learn about #PerturbSeq data but not sure where to start? working on the #VirtualCellChallenge? we're pleased to share differential expression summary statistic tables from five perturb-seq atlases collected by @josephmreplogle! (link below)
1/6 Excited to share our latest preprint: "MORPH Predicts the Single-Cell Outcome of Genetic Perturbations Across Conditions and Data Modalities". 🔗 biorxiv.org/content/10.110… 🧵 👇 Here is what MORPH is in a nutshell!
🤝Excited to announce @ProjectBiomni × @AnthropicAI! AI agents are set to transform how biologists do everyday research. Thanks to this partnership, the platform is now free for scientists worldwide: biomni.stanford.edu Learn more: anthropic.com/customers/biom…
The cells in our bodies constantly acquire mutations. But what are the patterns of mutations across tissues? How do mutations in normal cells lead to disease? These and other questions we will tackle within the SMaHT Network, now described in @Nature nature.com/articles/s4158…
1/ 🧵Can protein language models reason about evolution, not just model it? We built a 24 M-param PLM, Phyla, that reconstructs phylogenetic trees better than existing PLMs. Details & links in 2/👇
pgBoost, led by @elizabethdorans , is now out in @NatureGenet : highlights the utility of genetic effects on gene expression (eQTLs) to benchmark and unify multiome peak-gene links and genomic distance. nature.com/articles/s4158…
We are excited to share our latest preprint, as part of ADSP Functional Genomics Consortium FunGen-xQTL project, on a method called ColocBoost for multi-trait colocalization analysis. Mentored by @gaotwang @kanishkadey
Introducing ColocBoost - a new multi-phenotype xQTL and GWAS colocalization method, that can handle multiple causal variants and scale to 100 phenotypes, led by @cao_xuewei , and co-mentored with @gaotwang . medrxiv.org/content/10.110…
Introducing ColocBoost - a new multi-phenotype xQTL and GWAS colocalization method, that can handle multiple causal variants and scale to 100 phenotypes, led by @cao_xuewei , and co-mentored with @gaotwang . medrxiv.org/content/10.110…
Excited to announce our preprint describing SUPERB-SEQ 🦸, a new method to measure Cas9 edits and their effects on gene expression in single cells. Developed by @micklorenzini, myself, and @GrahamMcVicker biorxiv.org/content/10.110…
Excited to share our latest preprint, as part of ENCODE4, on a consensus variant-to-function (cV2F) score for functionally prioritizing variants for complex disease - led by Tabassum Fabiha and co-mentored with Alkes Price. biorxiv.org/content/10.110…
Analysis of single-cell CRISPR perturbations indicates that enhancers predominantly act multiplicative cell.com/cell-genomics/…
How are promoter interactions coordinated during cell differentiation? In our first paper, we investigate the temporal relationship between genome folding, transcription and histone modifications during ESC differentiation. (1)
Online Now: Transient promoter interactions modulate developmental gene activation dlvr.it/TFrfM2
Tri-i CBM held our annual retreat 10/27-28. We welcomed the new first years, had wonderful talks and discussions, and enjoyed rejuvenating group bonding time⭐️! Application portals are now open for perspective applicants, and we invite you to join our CBM family!