Michael Steidel
@michaelsteidel1
#Cellzome #TeamMassSpec opinions are my own
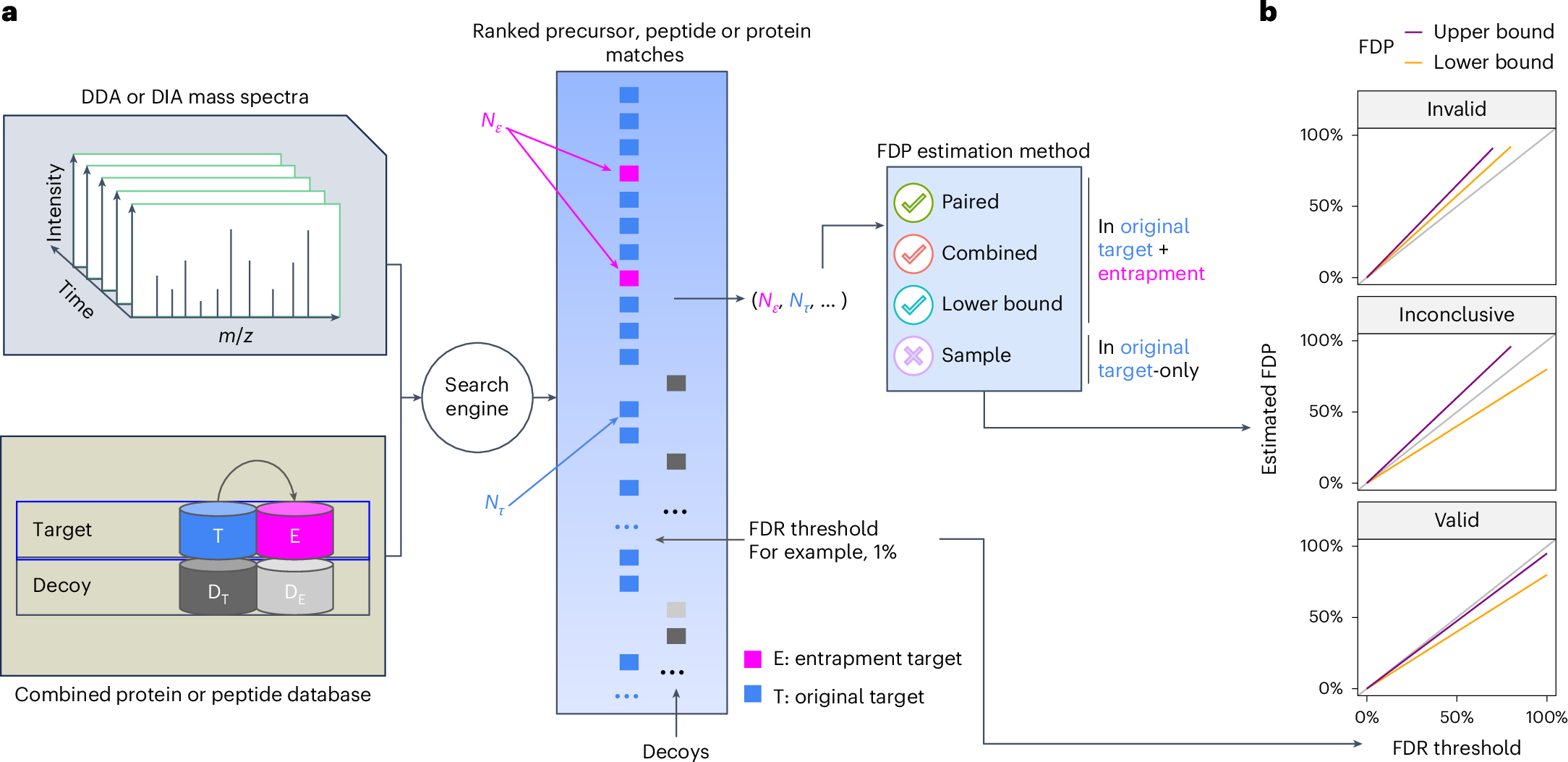
#TeamMassSpec, Any opinions on why not generally adding the relatively small yeast proteome to the anyway large human search space (*.fasta) as an internal FDR quality control? nature.com/articles/s4159…
My 2 cents on the current #TeamMassSpec #proteomics tools discussion: I fully support charging industry and keeping some modules closed source. Many free open-source tools lose relevance post-publication. In contrast, Alexey’s team stands out for maintaining their software at an…
Astral Zoom hits >7,000 protein groups & 67,000 precursors — on a 500 SPD EvoSep ENO run. biorxiv.org/content/10.110…

Protein digestion in 200 nL on solid-phase tips: SPEC enables low-input proteomics—even with SDS/SDC at high levels. From FFPE and muscle to high throughput plasma. SPEC is a universal and Evosep-compatible protocol. biorxiv.org/content/10.110…
biorxiv.org/content/10.110… timePlex enables time-domain sample multiplexing in LC-MS — boosting proteomics throughput up to 9× with no labels and minimal compromise in quant accuracy. Combine with plexDIA for 27 samples/run.
DIA-NN support for PAMAF (MOBILion): In proteomics, can we direct all peptide ions to fragmentation, therefore maximising sensitivity? We have previously introduced Slice-PASEF as a method of choice for single-cell level sample amounts. But what is the full potential of this…
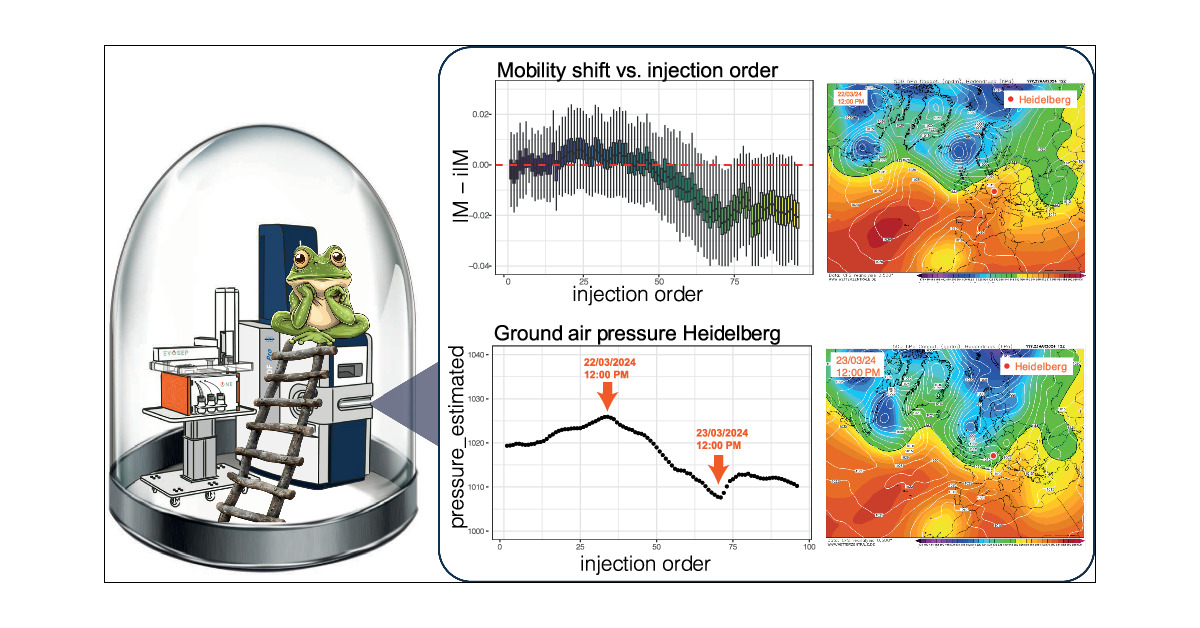
Weather’s going wild, and now your mass spec data’s a mess too? Coindidence? Nope! We reveal how weather-driven air pressure fluctuations impact diaPASEF-based high troughput proteomics - and how to fix it! pubs.acs.org/doi/10.1021/ac…
Hi #TeamMassSpec, Is it expected that diaPASEF file size triples from Pro2 to Ultra2? (From 2 GB to 6 GB, 60 SPD) ...
Hey #TeamMassSpec, I’m just now realizing that many of you seem to be gradually moving away from X. What is your preferred alternative? I gave up on my Mastodon attempt last year. Bluesky seems to have gained quite a bit of traction lately..
DIA-NN 2.0 release is almost ready, will come with some big news. One is scanning methods support, at the moment doing some tuning of the algorithm using Synchro-PASEF data by @labs_mann :) Turns out, 'Q1 information' is really helpful for gaining peptidoform confidence. Likely…
Our #diaTracer manuscript is out! 10 years ago we established the concept of library-free, direct DIA analysis with DIA-Umpire. DiaTracer now makes it possible to analyze any diaPASEF data, including PTM, semi-tryptic, nonspecific, and even open searches! nature.com/articles/s4146…
Hi #TeamMassSpec, Is there anyone having a function at hand to apply RT-dependent normalization of precursor quantities in the DIA-NN report. Would like to do this in a sample-group dependent fashion.
DIA-NN 1.9.2 is out! There are some major improvements, please check the release notes github.com/vdemichev/DiaN…