
Alexey Nesvizhskii
@nesvilab
Godfrey D. Stobbe Professor of Bioinformatics at U of Michigan. Trained as a theoretical physicist, now focusing on proteomics and proteogenomics.
Friends, #FragPipe 22 has been released, and it's a big update! diaTracer enables spectrum-centric analysis of diaPASEF data. Skyline integration. Koina server for more deep-learning prediction options. DDA+ mode for ddaPASEF. DIA glycoproteomics and new chemoproteomics workflows

Spent a great day in Barcelona at the EMBO Targeted Proteomics course hosted by @sabidolab, talking about our DIA tools and helping his team with a hands-on tutorial on FragPipe, FragPipe-Analyst, and Skyline integration, from installation and mzML files to pathway-level results.


Integrating Alternative Fragmentation Techniques into Standard LC-MS Workflows Using a Single Deep Learning Model Enhances Proteome ... biorxiv.org/content/10.110… #biorxiv_sysbio
The Nesvizhskii lab has 9 members attending #ASMS2025! 9 posters, contribution to 4 evening workshops, and one Bioinformatics Hub on #FragPipe. Plus multiple collaborative posters with other groups. See you in Baltimore!

See Kasia Kulej #USHUPO2025 poster on Decoding tumor-specific proteomes with integrative proteogenomics using ProteomeGenerator3 (plus comparison of DIA-NN, Spectronaut and MSFragger)
DDA is still great for many applications, and #MSFragger-DDA+ improves peptide identification rates via full isolation window search. Huge boosts in IDs, including Astral DDA! Fully integrated in #FragPipe, simply annotate your DDA files as DDA+ and RUN. nature.com/articles/s4146…

Tim Griffin, @nesvilab and Bing Zhang, will have a panel discussion on 'MS-Based #Immunopeptidomics: Challenges and Opportunities in Immuno-Oncology Research' at the #USHUPO2025 conference on 23rd Feb. ushupoconference.org/agenda#1-17403…
Our #diaTracer manuscript is out! 10 years ago we established the concept of library-free, direct DIA analysis with DIA-Umpire. DiaTracer now makes it possible to analyze any diaPASEF data, including PTM, semi-tryptic, nonspecific, and even open searches! nature.com/articles/s4146…
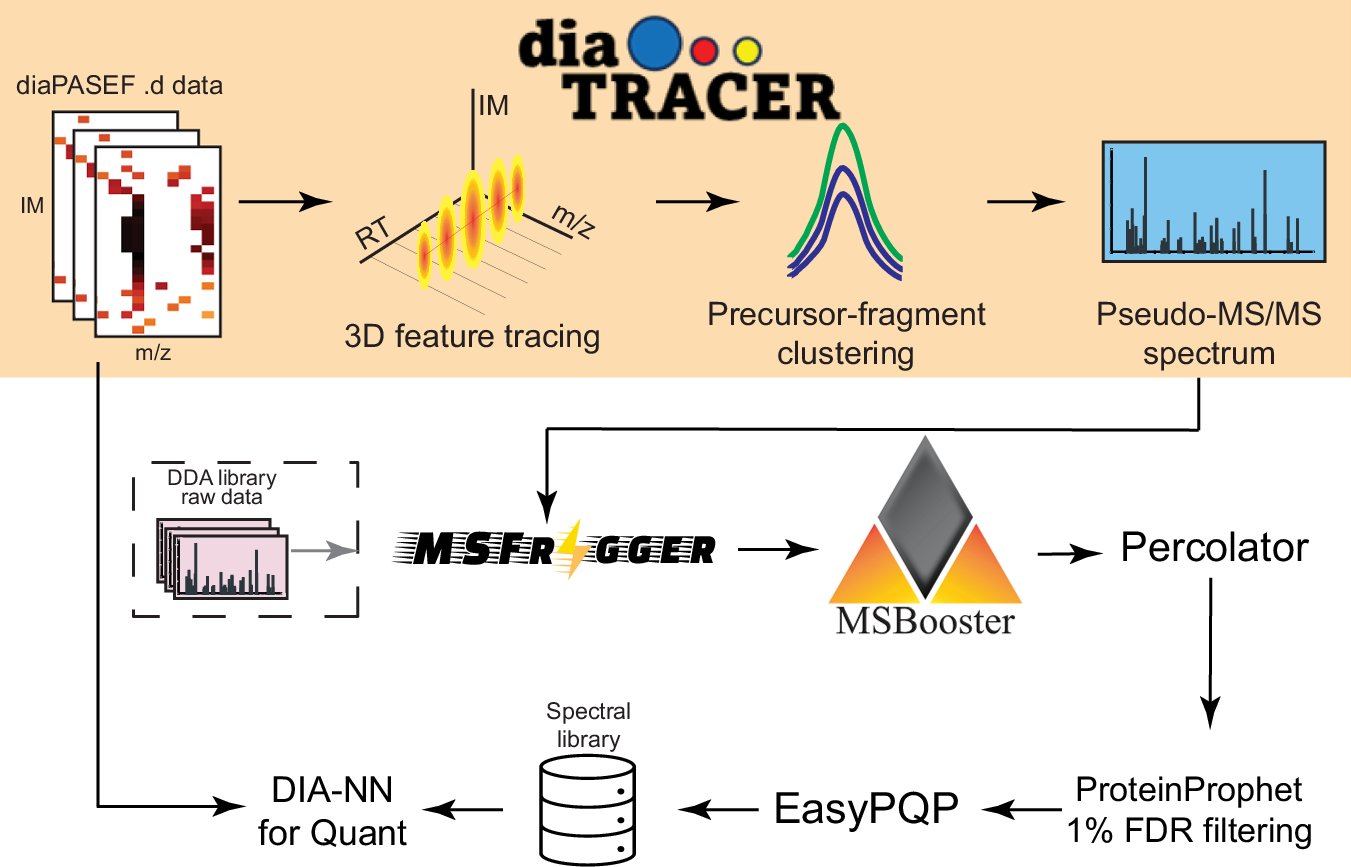
.@KaiPeiLi @nesvilab @fcyucn present diaTracer, a spectrum-centric tool for diaPASEF data, enabling direct peptide identification and quantification without spectral libraries. #BiotechNatureComms nature.com/articles/s4146…
Big news in QUANTITATIVE proteomics! Researchers from @UMich & @MonashUni have launched FragPipe-Analyst: a user-friendly, web-based tool to complement the popular FragPipe platform. Upload files, analyze, & visualize data with ease. #Precisionmedicine proteomics.cancer.gov/news_and_annou…
Great to see them apply the proteome-wide profiling method for electrophile reactivity and selectivity that we developed together with @nesvilab in Fragpipe in this exciting system. chemrxiv.org/engage/chemrxi…
Ok, my friends led, I followed. A few clicks, and here we go. Loaded the first photo I found on the disk (note to myself to change it later, way too serious). But 0 followers!! Now I know the feel of social isolation! Help me out there. I will not be leaving X entirely, not yet.

Very nice paper in Science, showing that N-glycosylation can also function as a degradation signal, similar to Ubiq: science.org/doi/10.1126/sc…. Glad to see the authors used our #FragPipe/#MSFragger-Glyco “Glyco-N-LFQ” workflow for quantitative glycoproteomics data analysis.
Assessment of Data-Independent Acquisition Mass Spectrometry (DIA-MS) for the Identification of Single Amino Acid Variants mdpi.com/3029520. Glad to see #FragPipe/#MSFragger showing the most conservative and effective performance (lowest false discovery match ratio (FDMR))
If your presentation falls on Halloween, do it in style! Here is Yi (Leo) Hsiao, Bioinformatics Ph.D. student in the lab, presenting his FragPipe-Analyst tool at today's "Tools and Technology" seminar series at the University of Michigan. #FragPipe medschool.umich.edu/events/tools-t…

Another great collaboration with the Keri Backus' lab, where we implemented a two stage search in #FragPipe to identify gain-of-cys mutations and variants proximal to reference cysteines. Two-stage searches can be useful for many applications, and they ensure group-specific FDR.
Excited to share our chemoproteogenomics story now out in its final form in @NatureComms. Congrats to first author Heta Desai @heta_desai17! nature.com/articles/s4146…
Kevin Yang from the @nesvilab closing the data analytics session by showing the power of MSFragger for scp-MS - incredible what free, academic software can do! #iSCMS2024
Nice protocol manuscript on low-input proteomics, with a description of the parameters in the LFQ-MBR workflow of #FragPipe the authors used.
Nice work by Matt @mrwaas and rest of the team to put this protocol together Protocol for generating high-fidelity proteomic profiles using DROPPS sciencedirect.com/science/articl…
I had to shorten my trip to Dresden for #HUPO2024 and have a room for Friday and Saturday night, and the following Wednesday night, that will sit empty. They require 14 day cancelation notice. If anyone in a desperate need for a place to stay on those nights, email me.
Come to my poster at HUPO on Tuesday! Direct spectrum-centric analysis of diaPASEF data using diaTracer and #FragPipe. See how well it works for spatial proteomics, CSF, plasma, HLA, PTMs and more! Updated preprint: biorxiv.org/content/10.110…. With co-authors Kai, Fengchao, Guo Ci.

Rethink the analysis of DDA data! Blurring the boundary between DDA, WWA, and DIA, #MSFragger-DDA+ Enhances Peptide Identification Sensitivity with Full Isolation Window Search. Fully integrated in #FragPipe, simply annotate your DDA files as DDA+ and RUN. biorxiv.org/content/10.110…
