
Leo Speidel
@leo_speidel
Statistical and Population Genetics | RIKEN, iTHEMS (Japan) | 🇯🇵🇩🇪🇬🇧 |
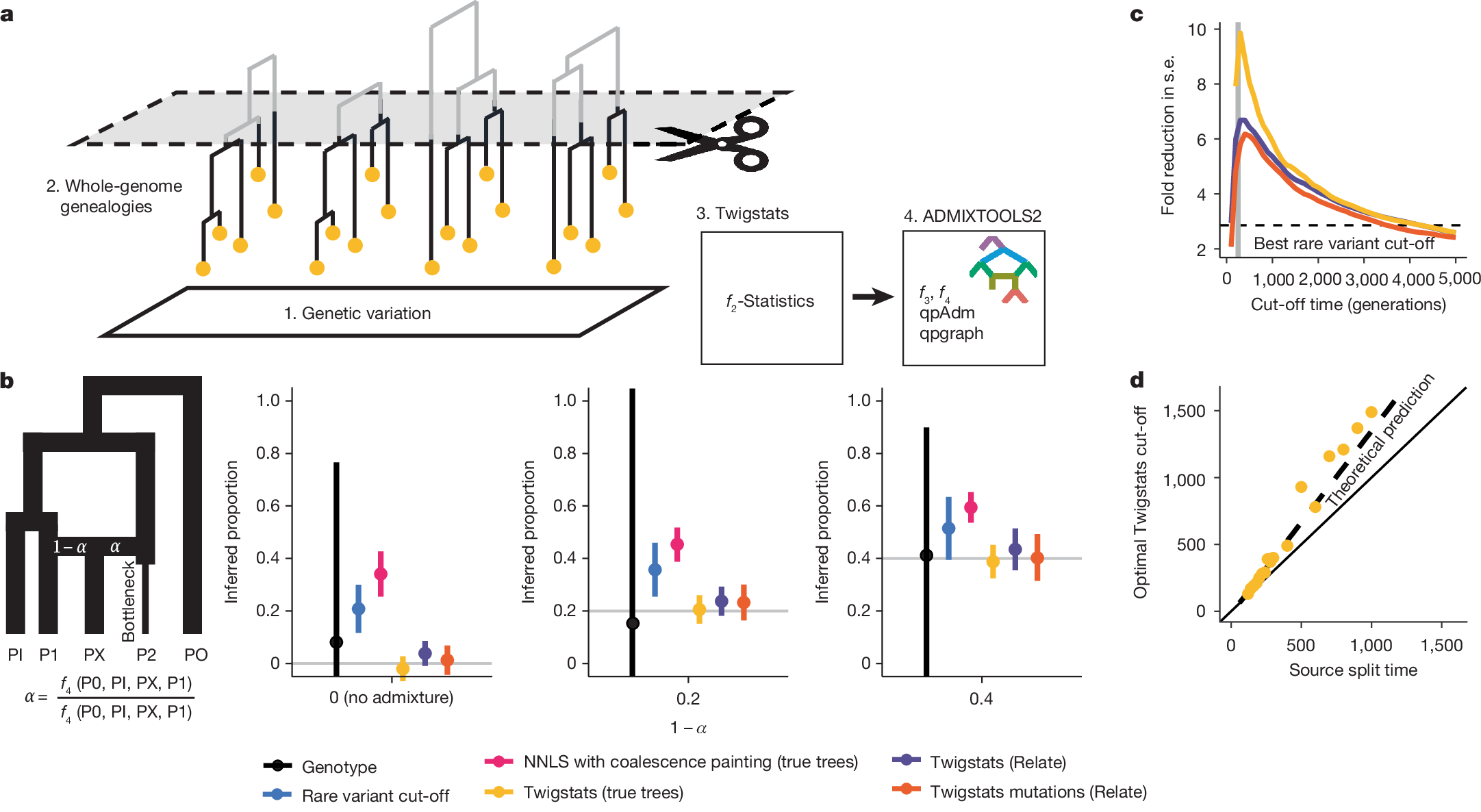
Happy New Year! Our Twigstats paper is now out! nature.com/articles/s4158…
Postdoc position in my group in Tokyo! Please get in touch if you are interested. And happy to discuss projects - ranging from developing new methods to analysis of new genomes that we are now sequencing in the lab. riken.jp/en/careers/res… Lab page: speidellab.github.io

(1/6) Excited to share our manuscript describing Quickdraws, a novel algorithm and software to perform GWAS that increases association power while keeping computational costs low. More details below (thread). Link to article: nature.com/articles/s4158…
インタビュー【MEET a rising star at RIKEN】 理研で活躍する若手研究者が理研の魅力や研究環境、日々の暮らしを語ります。第1回はシュパイデル玲雄(@leo_speidel)理研ECLユニットリーダー。数学・統計学を用いてヒトの進化・歴史を追求しています。 📹 youtu.be/T-4VhIbllow
New from @wwTianyiww in Science science.org/doi/10.1126/sc…
Our new paper is out in @ScienceMagazine! By exploring the rich genetic diversity of Brazil, we show how fine-scale genomic analyses reveal that this diversity, rooted in Indigenous ancestry and centuries of complex demographic history, plays a key role in population health.
I have a 2 year postdoc position available on our 'COREX: from correlations to explanations' grant: cordis.europa.eu/project/id/951…. See: ucl.ac.uk/work-at-ucl/se…... working at the intersection of archaeology, computational modelling and stats. Starts Nov 2025.
これまで統合生物考古学セミナーや進化学会夏の学校などで扱ってきました内容を中心に,「ゲノム多様性解析」として森北出版から出版予定です.多くの方のご協力を仰ぎました. 類似の日本語教科書はないと思いますので,ご興味のある方はぜひ手にお取りください. morikita.co.jp/books/mid/0261…
Applications are now open for the #EMBOpopgen course - deadline 24th Feb! All information here: meetings.embo.org/event/25-pop-g…
Our papers, out today in @nature, show how ancient DNA from the Eneolithic and Bronze Age steppe points to a North Pontic origin of the Indo-European language family and a Caucasus-Lower Volga (CLV) origin of Indo-Anatolian (inclusive of the now extinct Anatolian languages). 1/
Congratulations Sile! And really fascinating & clean result seeing conservation of effect sizes across human groups using local ancestry in admixed people!
Excited to see my project at @UniofOxford finally published at @NatureGenet ! Huge thanks to my co-authors @simon_r_myers @danjlawson @marchini @g_hellenthal @linoafferreira @ShiSinan , and the reviewers/editors. Read the publication here: nature.com/articles/s4158… 🚀🎉
Excited to see my project at @UniofOxford finally published at @NatureGenet ! Huge thanks to my co-authors @simon_r_myers @danjlawson @marchini @g_hellenthal @linoafferreira @ShiSinan , and the reviewers/editors. Read the publication here: nature.com/articles/s4158… 🚀🎉
Excited to share our preprint "A putative ecocline in Klebsiella pneumoniae" and super-interested to get your feedback. Really want to use the hive mind to get to the bottom of this phenomenon ASAP. biorxiv.org/content/10.110…
Modern GWAS can identify 1000s of hits but it can be hard to turn this into biological insight. What key cellular functions link genetic variation to disease? I'm excited to present our new work combining associations + Perturb-seq to build interpretable causal graphs! A🧵
One week to the abstract submission deadline for SMBE 2025, Beijing 20-24 July! smbe2025.scimeeting.cn @ana_ignatieva and I are organising a symposium on popgen through time: using ARGs, aDNA, or otherwise to understand the evolutionary processes that shape genomes through time.

A genealogy-based approach for revealing ancestry-specific structures in admixed populations biorxiv.org/cgi/content/sh… #biorxiv_genetic
Delighted to see our new paper out in Nature nature.com/articles/s4158…
Our new paper is out! Congrats to the whole team, especially the staff and students @bournemouthuni who have been excavating this incredible site for 15 years (with much more left to uncover!) Widespread matrilocality in Iron Age Britain - Up Na Mná :) rdcu.be/d6mb0
Very happy to share the results of my @MSCActions project done at @UGI_at_UCL with @mt_genes and @pontus_skoglund from @TheCrick. We used ancient DNA to reveal remarkably high genetic diversity in the region of modern-day @Ukraine over the last 3,500 years until ~500 years ago.
2025 is off to a great start. Illustrated @nature's first cover of the new year!🐲🥳 Fascinating new research led by @leo_speidel and @pontus_skoglund
Kicking off the new year, our paper by @leo_speidel et al. is out describing what to me is a breakthrough in ancient genomics. By using genealogies to study ancestry we can get much more resolution to study finer-scale history. We call this method Twigstats.
My friend Pontus Skoglund and his colleagues have JUST published this incredible new piece of research into Viking-era migrations - using ancient DNA and a brand new method called TWIGSTATS. This epic piece of research is matched by an epic cover of first 2025 issue of Nature!