Hope Tanudisastro
@htanudisastro
MD-PhD student @Sydney_Uni and @GarvanInstitute Centre for Population Genomics | studying STRs in the @dgmacarthur lab
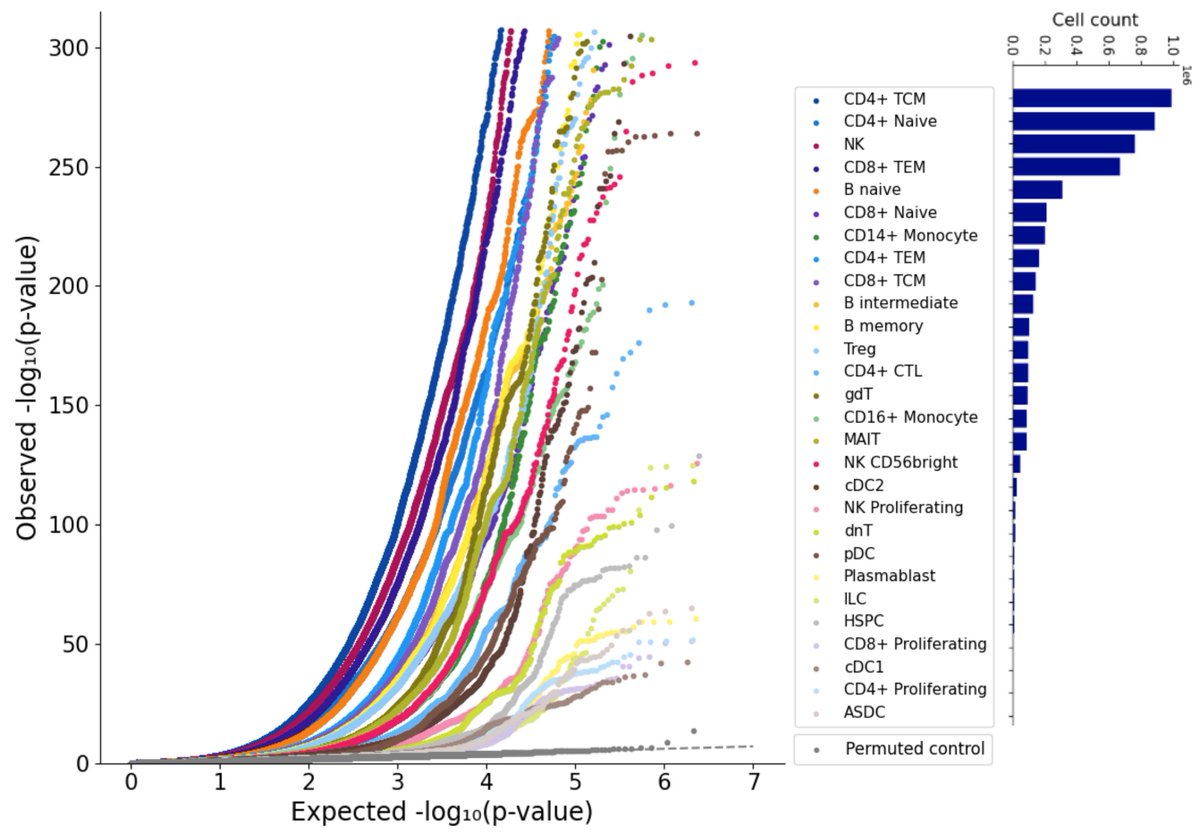
📢 New preprint alert: We’ve just published a deep dive into the role of tandem repeats (#TRs) in single-cell gene expression across the immune system, using #WGS and #scRNA-seq data from 1,790 individuals and over 5 million blood cells! 🧬 🧵👇 #repeats #SingleCell 1/9
In @NatureGenet we report that base editing of trinucleotide repeats (TNRs) reduces somatic repeat expansions in Huntington’s disease (HD) and Friedreich’s ataxia (FRDA)—in patient-derived cells and in vivo—a collaboration with the Mouro Pinto lab. drive.google.com/file/d/1on7kdy… 1/14
It was great to be able to contribute the EP400 finding to this paper together with @Gianina_Natoli and @ChiaraFolland , flagging a novel CAG repeat locus in the EP400 gene as a likely (though extremely rare) cause of spinocerebellar ataxia when expanded beyond 55 repeats.…
Detailed tandem repeat allele profiling in 1,027 long-read genomes reveals genome-wide patterns of pathogenicity biorxiv.org/cgi/content/sh… #biorxiv_genomic
Looking forward to attending #ozsinglecell2025. I’ll be sharing some findings on tandem repeats #TRs in single cell contexts using TenK10K Phase 1 on Thursday at 11:55.
Thrilled to share my latest work on somatic instability in X-linked Dystonia Parkinsonism. This is part of a large effort in uncover the genetic basis driving somatic instability in repeat expansion diseases biorxiv.org/content/10.110…
In a medical milestone, a customized base editor was developed, characterized in human and mouse cells, tested in mice, studied for safety in non-human primates, cleared by @US_FDA for clinical trial use, manufactured as a complex with an LNP, and dosed into a baby with a severe,…
Nice! Environmentally responsive eQTLs are enriched for being more distal and for constrained genes relative to conventional QTLs. biorxiv.org/content/10.110…
Complete view of de novo mutation rate across the genome estimated from a four-generation pedigree using five complementary short and long read sequencing technologies. The authors estimate 98–206 DNMs per transmission: - 74.5 SNVs - 7.4 tandem repeats - 65.3 structural…
Having your phone on your desk or in your pocket undercuts cognitive performance - even if you don't use it. Put it in another room to maximize your working memory and fluid intelligence.
Our vision for Precision Omics Initiative Sweden (PROMISE) is now out in Nature Medicine! By boosting data-driven precision omics in Sweden, we can not only empower research but also integrate it with healthcare in a new way, with an impact in 🇸🇪🌎. 🧵: rdcu.be/egmXY
1/10 Excited to share our latest - the first whole-body map of both DNA methylation and 3D genome at single-cell resolution.
🚨 Our latest GWAS of heart failure subtypes is now out in Nature Genetics! rdcu.be/eb9O0 A massive global collaboration from HERMES Consortium involving >40 studies with >150,000 heart failure cases. @HERMESgenetics @tomlumbers 🧵 highlighting key findings 1/
The TenK10K Phase 1 dataset is out! A mammoth effort to generate and analyze paired WGS + scRNA-seq at scale - already yielding exciting insights, with many more to come.
📢 new preprint alert: So so excited to share our analysis on the impact of common and rare variants on single-cell gene expression in blood, using WGS and scRNA-seq data from nearly 2,000 individuals and 5.4m cells as part of TenK10K phase 1 🧬medrxiv.org/content/10.110… 🧵👇 (1/n)
📣Excited to share my last postdoc paper with @soumya_boston on eQTL mechanisms depending on where the RNA is in the cell! @BrighamResearch @broadinstitute TL;DR:Early RNA eQTL variants in the nucleus and late RNA eQTL variants in the cytosol have distinct molecular mechanism🧵👇
Early and late RNA eQTL are driven by different genetic mechanisms biorxiv.org/content/10.110… #biorxiv_genomic
1/ Many bioinformatics students don’t know much about NGS pre-processing. But trust me, understanding the raw data is essential. Here’s why.
Only 2 weeks left to get legislation through before the next election in Australia to protect anyone who engages with genetic and genomic research from having to worry about it impacting their insurance. Please urge your local MP to make this happen abc.net.au/news/2025-01-2…
What if single-cell analysis of 1,000 samples wasn’t just imaginable—but affordable? Imagine analyzing 1,000 samples at the single-cell level, each containing ~25,000 cells or nuclei. That’s ~25 million cells in one experiment!. Until now, the costs of such an experiment were…
The abstract deadline for "Mutations in Time and Space 2025" is in a week! Please register here: broadinstitute.swoogo.com/mits2025
Profiling tandem repeats variations in the population using 1027 long read genomes from All of Us cohort. All the samples are from individuals of African and African American ancestries. I remember hearing about this dataset first time at ASHG 2022. Great to see the data out…
Our new Nature Genetics paper on human segmental duplications (SDs) provides a pangenome perspective of SDs, new potential protein-coding genes, and greater complexity in human variation with implications for disease and evolution. rdcu.be/d5AD9
Come and join the lab! we have a position available to work on the analysis of short and long-read DNA and RNA sequencing from patients with repeat expansion diseases jobs.ac.uk/job/DLG312/bio… #RepeatExpansionDisorders #RepeatExpansions #C9orf72