Robert K. Bradley
@bradleybio
#RNA and data science for #cancer therapy @fredhutch @uwgenome. Retweets are not endorsements.
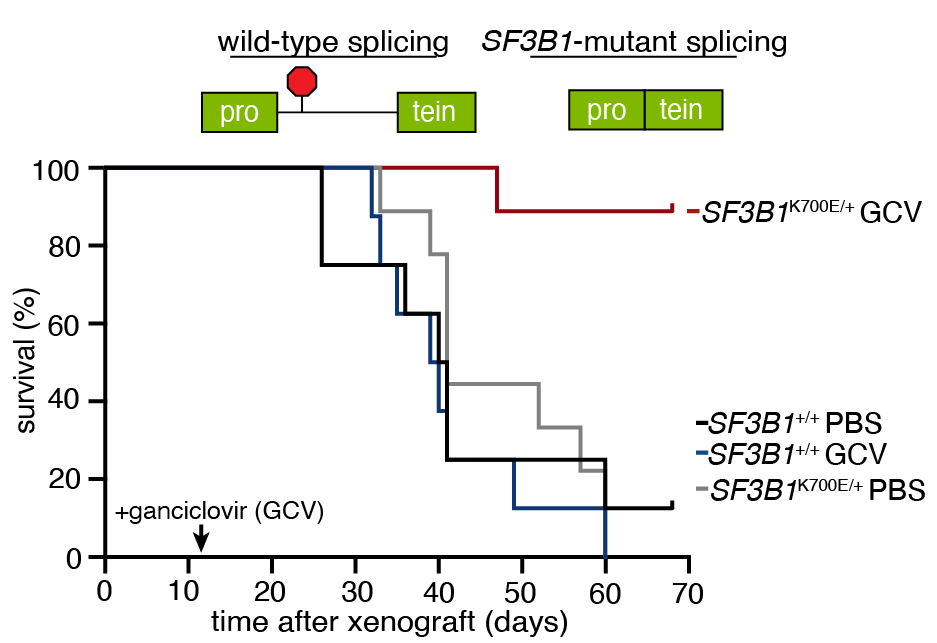
I'm excited share our "synthetic intron" technology for selective expression of therapeutic proteins in cancers, just out in @NatureBiotech. This was an amazing collaboration with @AbdelWahablab led by @khrystyna_north and @SBenbarche nature.com/articles/s4158…
Check out this very exciting study! So glad to see @satijalab focusing on RNA processing. Hoping for lots more insights to come!
New preprint w/@anshulkundaje introducing CPA-Perturb-seq! We systematically perturb regulators of cleavage and polyadenylation, and explore post-transcriptional changes at single-cell resolution. Led by @mh_kowalski @harm__w and @jjohlin (🧵) biorxiv.org/content/10.110…
Happy to share a new paper from work in @bradleybio lab demonstrating that APC is an RNA-binding protein that regulates poly(A) site selection. genomebiology.biomedcentral.com/articles/10.11…
I'm excited to share our study demonstrating that alternative polyadenylation regulates tumor growth in melanoma, just out in @NatureComms and led by @a_gabel2! nature.com/articles/s4146…
Are you an early career scientist who wants to pursue a research project in the lab of a Fred Hutch Computational Biologist mentor? If yes, then the #MahanFellowship is for you. Apply by Dec. 1, 2023! Learn more: apply.interfolio.com/130340
I'm excited to share PRIMeD! It's an awesome opportunity for recent PhD or MD graduates to visit @FredHutch, present your work, and learn about #immunology, #oncology and #DataScience! Deadline is July 15; please apply! fredhutch.org/en/events/prim…
Excited to share PRIMeD! Calling all PhD (or equivalent) students within 2 years of graduation or who have graduated in the last year: Join us for an opportunity to visit @FredHutch and learn about #immunology, #oncology and #DataScience! fredhutch.org/en/events/prim…
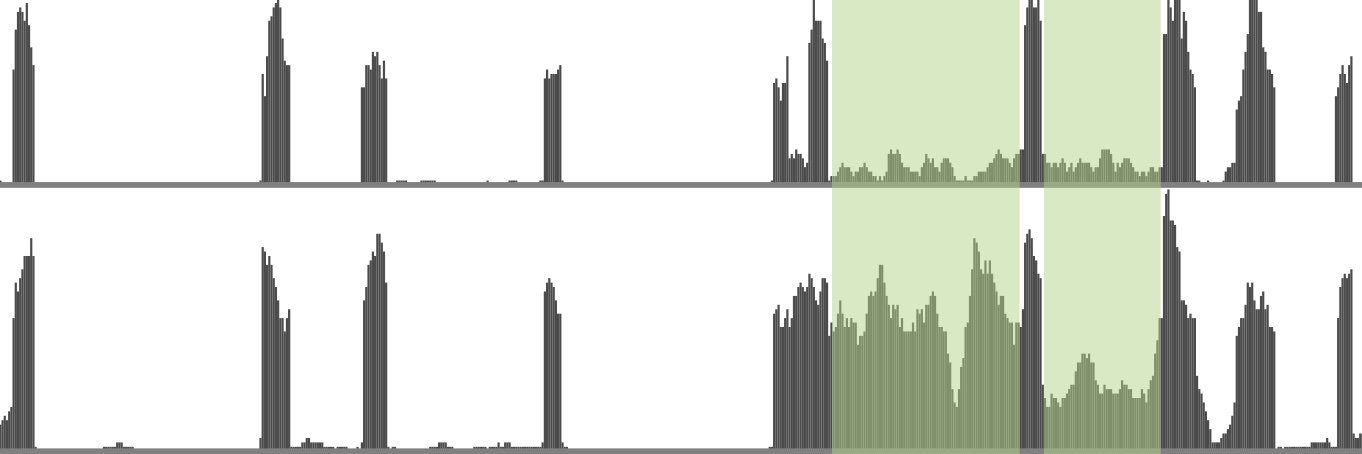
@emma_hoppe @bradleybio from @fredhutch report that recursive splicing occurs across a broad range of human introns and that these sites are frequently conserved and influence alternative splicing. bit.ly/3nCrgt8
We're hiring a research technician to join our team! Please apply if you're interested in manipulating #RNA processing to create new #cancer therapies. careers-fhcrc.icims.com/jobs/25079/res…
Excited to co-organize with @eirinipapapetr1 the next Workshop on Splicing Factor Mutations and RNA biology in cancer here at Sinai. Registration is open (deadline April 3). Looking forward to seeing you in NYC. #RNAbiology #RNA #Splicing #Cancer
NEW content online! RNA splicing dysregulation and the hallmarks of cancer dlvr.it/Sggn8w
I'm pleased to share a new review article on functional connections between RNA splicing and the hallmarks of #cancer that I wrote with @OlgaAnczukow, just out in @NatureRevCancer nature.com/articles/s4156…
Fascinating study from @SchragaSchwartz identifying strong and causal links between splicing and m6A deposition.
Our work, demystifying m6A deposition, is out! m6A is not selectively installed. It is installed by default, at all eligible consensus motifs, except in the vicinity of splice junctions, where it is excluded by the exon junction complex authors.elsevier.com/a/1gMUY3vVUPK9…
Today is the last day to apply to our open #DataScience faculty position @fredhutch! apply.interfolio.com/115753