
Heidi Rehm
@HeidiRehm
Genomic medicine researcher; chief genomics officer at MGH; clinical medical officer at Broad Clinical Labs @broadinstitute; Views are my own.
What a delight to be kicking off our inaugural @OurDNA_program symposium in Melbourne with a keynote from @HeidiRehm discussing global collaborative networks underpinning genomic medicine. Thanks to @AusGenomics and @GA4GH for making this day possible!
The #ClinGen expert curation ecosystem enables the global dissemination of evidence-based #genomics support resources bit.ly/3Vg9Hgj #clincalgenetics #datasharing @splon @HeidiRehm @ClinGenResource
We are delighted to announce this year's call for applications for the NHGRI-funded MGB T32 Postdoctoral Training Program in Precision and Genomic Medicine buff.ly/414Xajk Please RT or share w/ those who may be interested.

🔥 Fireside Chat happening now! We're on Zoom with Bartha Knoppers of McGill University and Gerardo Jiménez-Sánchez of Genómica Médica in conversation about Dr. Jiménez-Sánchez career and contributions to the field of genomics. Please join us! Join here: hubs.li/Q02ZlnHd0
I know many of you have been awaiting us launching transcript expression data in gnomAD. We were waiting for the GTEx v10 release which is now out so we are finally able to launch this. Enjoy!!
Proportion expressed across transcripts (pext), using GTEx v10, is now available on #gnomAD v4!
Just started using Buffer to simultaneously post on @bsky.app, LinkedIn, and X/Twitter as I make my gradual shift away from the toxic and biased world of X....
Forthcoming guidance will recommend labs report VUS subclasses. We share experience of 4 labs including rates of reclassification of VUS subclasses. By highlighting VUS-high and downplaying VUS-low, this will be game-changing for dx genetic testing. buff.ly/3Cpx5RL

📝The Health Data Sharing, Privacy, and Regulatory Forum — formerly known as the GDPR & International Health Data Sharing Forum — publishes Policy Briefs on emerging and relevant health data sharing policy topics. Read this month's policy brief: bit.ly/4fDhlJn
Great stuff from @AnneOtation and @VGaneshMDPhD and others!
Deletions in a gene (𝘊𝘏𝘈𝘚𝘌𝘙𝘙) that encodes a long noncoding RNA cause severe developmental delay and an increase in expression of a neighboring gene, 𝘊𝘏𝘋2. Read the Brief Report: nej.md/48gS1WY
See our paper in GIM here. If you publish on variants, PLEASE use variantvalidator.org, or even better, just submit them to ClinVar before article submission (you can request a 6 month embargo), to both validate your variant naming AND make them findable by all!
GIM's Senior Technical Editor Peter Freeman collaborated with other experts on "Standardizing variant naming in literature with VariantValidator to increase diagnostic rates", now available in Nature Genetics bit.ly/3BF83On
It’s great to have these more refined allele frequencies from the African/AA population represented in gnomAD. Thank you team!!
I'm excited to announce that we have generated local ancestry informed allele frequencies for the inferred African/African American genetic ancestry group of gnomAD v4.0, live now on the browser! gnomad.broadinstitute.org/news/2024-10-l…
Available today in @GIMJournal, a special article from @ClinGenResource “The Clinical Genome Resource (ClinGen): Advancing genomic knowledge through global curation” sciencedirect.com/science/articl… (1/n)
I am pleased to announce the establishment of an African Bioinformatics Institute thanks to funding from Wellcome Trust and CZI. We seek stakeholder input to shape the ABI to meet the needs of the bioinformatics and user communities. Join us for engagement sessions to contribute.
The deadline to apply for a full time faculty position in Neurogenetics at the @CGM_MGH and @MGHNeurology is October 15th! Send your application in to be considered.
@CGM_MGH and @MGHNeurology are seeking applications for a full-time CGM faculty member with an assistant or associate professor of neurology appointment at @harvardmed! loom.ly/GbLy26E
Terrific #WMIF2024 panel discussion on 'Earliest Detection' chaired by @JimBrinkMD & D. Louis (@MassGenBrigham Radiology & Pathology Enterprise Chiefs, respectively) toward ambitious primary prevention across neurologic, CV, & genetic disease
Jasmeer Chhatwal and Yakeel Quiroz join a panel on Earliest Detection during #WMIF2024 @ytquiroz
A wonderful meeting with so many folks dedicated to advancing genomics through collaboration and open data sharing.
GA4GH 12th Plenary 🤩 🇦🇺: Melbourne, Australia and online 👥: 650 registrants 🌐: 44 countries 💼: 3 meetings, 65 sessions Big thanks to our co-host, @AusGenomics 🤝 🇸🇪: 13th Plenary will be in Uppsala, Sweden Read the full Plenary recap here: hubs.li/Q02QxcQt0
Welcome to GA4GH 12th Plenary! This week, we are gathering to advance responsible sharing of genomic and related health data, for the benefit of human health. Learn how you can participate this week, from wherever you are located: bit.ly/4e5Hlg1
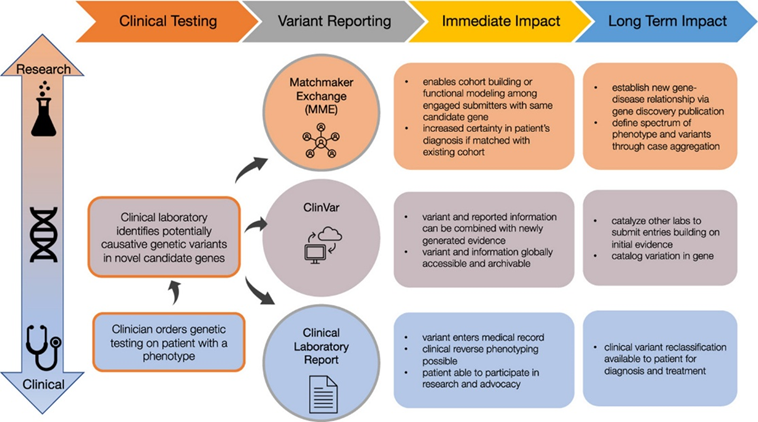
As part of @GREGoR_research , we are excited to announce new guidance to encourage clinical labs to report novel candidate genes during clinical testing. We give criteria for triaging, sharing, and reporting novel genes through various methods. authors.elsevier.com/c/1jcDA3vlFV4E…
GA4GH 12th Plenary is next month! 🎉 You can now register for virtual participation. In-person registration ends 2 September. #joinus Register here: bit.ly/3yRSRMB
Wonderful news!!! Congrats @aaronquinlan
When I started grad school in 2004, I never could have imagined I would write the following sentence. I am honored and thrilled to be the Chairperson of Human Genetics @UUtah! I look forward to future excellence w/ current & new scientists in our amazing department. Stay tuned!