Scott Coyle
@CellRaiser_
cells and proteins. molecular machines. Assistant Professor: @uwbiochem | Postdoc: @stanford @prakashlab | Ph.D.: @ucsf Wendell Lim @CDI_UCSF
Our first paper is out in @CellCellPress! Led by @born2raisecell, we present a programmable reaction-diffusion system that generates synthetic protein waves, oscillations, and patterns for spatiotemporal circuit design and FM data-encoding in human cells. authors.elsevier.com/a/1iN8ZL7PXmeeN
Excited to share a new paper from the super creative and talented @EdenChang_! Eden engineered droplet-forming sequences to act as regulatable glues for stitching microtubules together inside living cells 🧫🧪
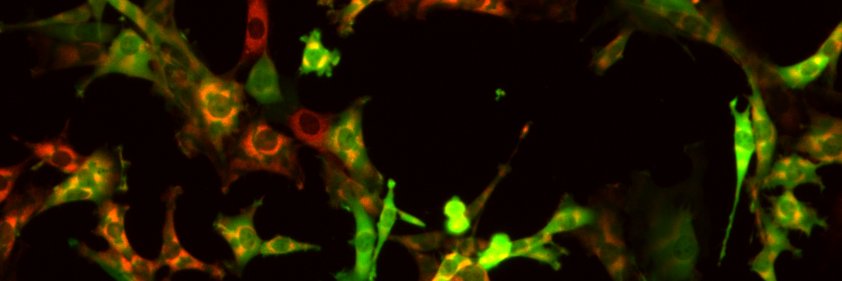
I'm thrilled to share my first project in @CellRaiser_ 🤩we found that MTs-assembly can be tuned at both MTBD and condensates. This project highlights my past(microtubule), present(SynBio), and future(LLPS): doi.org/10.1016/j.jbc.…
Thrilled to share our collaborative study led by Brent Aulston and @Musing_Naiad in my lab! 👇This work is a pivotal proof-of-concept in mice showing that in vivo #CRISPR editing in the brain has potential to treat Alzheimer’s disease. 🌟🧠 @NIGMS @somaticediting
Gene editing of APP C-terminus rescues neuropathology, ephys, behavior, and transcriptomic changes in #alzheimer knockin mice. Led by Brent Aulston, collaboration with labs of @sahakris @MarkZylka @MarceloWoodUCI Jon Audhya (Madison) #crispr #therapeutics biorxiv.org/content/10.110…
Some say hybrid comp/expt labs, where dry lab meets wet lab, are "at the beach". Hbt "the shore of Lake Mendota" ? I'm thrilled to join Biochemistry at UW-Madison! Starting 2025, the Wayment-Steele lab will unite AI & experiment to transform how we study biomolecular dynamics.
Also check out the great write-up by Eli Ramos and @Taylor_Kubota at Stanford Reports: news.stanford.edu/stories/2024/0…
👀Glad to see this story out on algae-animal symbiosis🤗and the role of photosynthesis☀️, a wonderful collaboration led by the amazing @NanesDania from @PhysPlanaria lab 🥳🥳 @carnegiescience @Stanford nature.com/articles/s4146…
enjoy the awesome experience, science, and people @kyle_flickinger and @JustWadeAMinute! super proud of & very excited for you both! way to rep metabolism & interdisciplinary work @uwmadisonipib @UWBiochem @Morgridge_Inst cc: @JudithSimcox @RutterLab @gsducker
On Monday (May 20th) starting at 9am in HSEB 1700, we look forward to welcoming 10 talented Graduate Student Rising Stars! Don't Miss it! @uofunuip @AnnaliseBond @Heankel_ @JustWadeAMinute @jaee_saucy @MarissaNT96 @kyle_flickinger @dumebiike @Sam_K_Zepeda @DucHuynh1393 @dacolon
Ingenious strategy to repurpose a bacterial E1-like enzyme to emulate ubiquitin-like cascades for ATP-driven protein ligation. Its like a battery pack for powering bioconjugation reactions 🔋!
On Monday (May 20th) starting at 9am in HSEB 1700, we look forward to welcoming 10 talented Graduate Student Rising Stars! Don't Miss it! @uofunuip @AnnaliseBond @Heankel_ @JustWadeAMinute @jaee_saucy @MarissaNT96 @kyle_flickinger @dumebiike @Sam_K_Zepeda @DucHuynh1393 @dacolon
Our paper is finally out in it's final form!
Online Now: The full spectrum of SLC22 OCT1 mutations illuminates the bridge between drug transporter biophysics and pharmacogenomics dlvr.it/T6VdRZ
At #ASGCT2024, we're showcasing advances in non-viral gene editing, CAR T cell therapies, & high-throughput screening. Explore our work on vision restoration, cancer therapy, and ethical challenges in the field. Join us to learn about the latest progress in #GeneTherapy research
Following our field's inspiring history of building synthetic receptors, here we share our lab's first attempt at a post-transcriptional one. Based on ADAR-editing and compatible with various inputs/outputs.
Post-Transcriptional Modular Synthetic Receptors ift.tt/Yu9qIdk #biorxiv_synbio
Happy to share that work led by grad students Zack Harmer and Jaron Thompson, with significant contributions from David Cole, is now out in the journal @ACSSynBio. This work developed out of a productive collaboration with the research groups of Zavala and Venturelli.
Imaging the entire course of tissue regeneration requires transparent animals, efficient transgenesis & microscopes that can image free-behaving animals. Here we present a toolkit and reveal dynamic cellular processes during regeneration of Macrostomum. biorxiv.org/content/10.110…
ASCB MEMBER NEWS Judith Kimble & Others Wins the 22nd Annual Wiley Prize in Biomedical Sciences Congrats to ASCB Member Judith Kimble & colleagues who won the 22nd annual Wiley Prize in Biomedical Sciences for their discovery of the stem cell niche. ow.ly/bhrX50Rpfhf
Excited to present PHAGEPACK for quantifying genome-wide which bacterial host factors impact phage infectivity. PHAGEPACK combines CRISPRi with phage packaging. Reveals phage-host interactions, evolution, & host resistance. Great work by @chuchitboonthav! biorxiv.org/content/10.110…
Congratulations to Assistant Professor Monica Neugebauer (@NeugMonicaElena), who is a 2024 recipient of the Shaw Early Career Research Award! The $200,000 award is sponsored by @GrMKEFdn. Learn more about Neugebauer's research: biochem.wisc.edu/2024/04/24/neu…
Incredibly honored to be selected as one of the 2024 @SchmidtFellows! Excited to join the SSF community and collaborate with so many inspiring individuals. Looking forward to this journey ahead!
We’re so excited to announce our 2024 Schmidt Science Fellows. 32 exceptional early career scientists will join our global community to pursue interdisciplinary research and help change how science is done. schmidtsciencefellows.org/news/eric-and-…
The regulatory capacity of dynamic systems is rich & fascinating. They can enable a common regulatory hub to differentially regulate diverse physiological processes. pubs.acs.org/doi/full/10.10…
We have a new preprint on optical patterning of Nodal signaling in zebrafish embryos: using light to sculpt embryonic development! biorxiv.org/content/10.110… Beautiful work led by @LordLab_Pitt and @h4rrymcnamara
We’re very excited to share the first preprint from my lab along with @h4rrymcnamara, @BillZJia, @AdamEzraCohen, and Alex Schier. We describe a set of methods for exerting spatial control over Nodal signaling in zebrafish development. biorxiv.org/content/10.110…
We made the cover of @CellSystems! Elementary landscapes for two fitness functions reveal that combining them creates a more rugged landscape. This highlights the demand on a DNA-binding domain to evolve affinity for distinct half-sites @Jackson_Lab tinyurl.com/yc2web9a
New Preprint out from the Vecchiarelli (@CellfOrganized) and Biteen (@JulieBiteenLab) Labs! "An experimental framework to assess biomolecular condensates in bacteria" biorxiv.org/content/10.110…