Thomas Macrina
@tmacrina
Zetta CEO, reconstructing connectomes
Excited to share a connectomics dataset of hippocampus CA3. Explore the dataset: pyr.ai Paper introducing the dataset and some of our discoveries: biorxiv.org/content/10.110…
Wouldn't it be great if we could not only image large connectomic volumes, but also completely reconstruct them? And if a whole mouse brain project didn't cost billions? With the PATHFINDER preprint (biorxiv.org/content/10.110…), we preview a future where it doesn't have to.
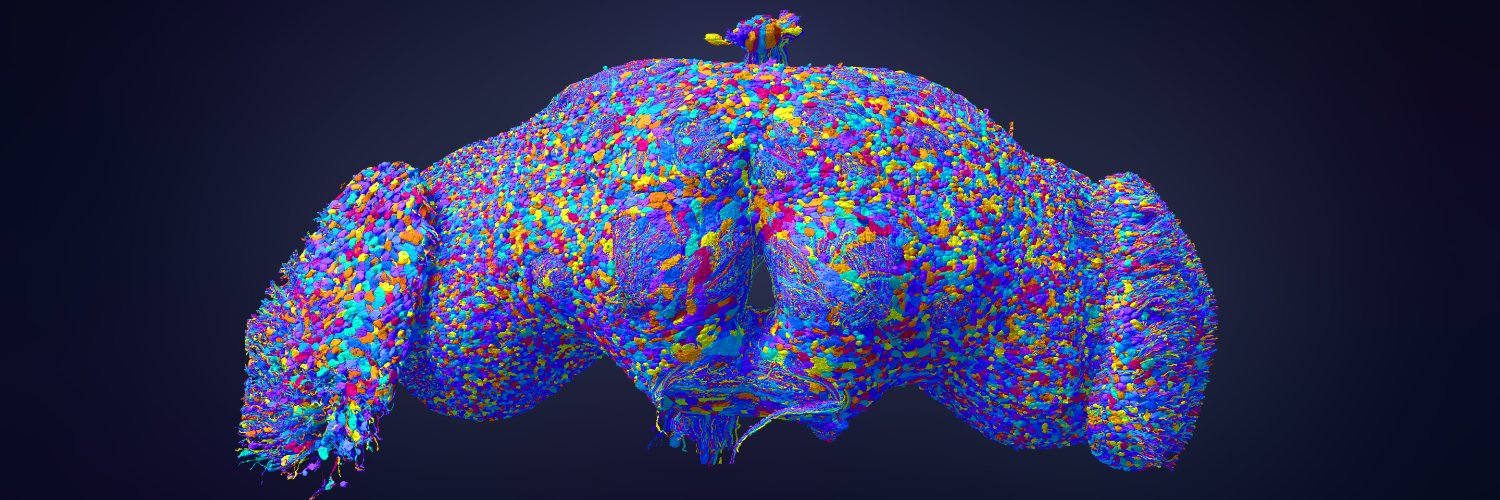
Still floors me that our MICrONS team opportunistically mapped the first fly brain as a side project, enabled by tools they had just developed for solving a much harder problem.
Shortly after that MICrONS milestone, I persuaded our team to shake off their exhaustion and apply the same image processing pipeline to the fly brain as a side project. This led to the automated reconstruction that become the basis of FlyWire. x.com/FlyWireNews/st…
As people that know me well can attest, I love a good mystery! 🔍 Fortunately for me, this work had twists both surprising and peculiar. 🧵
New Anthropic research: Tracing the thoughts of a large language model. We built a "microscope" to inspect what happens inside AI models and use it to understand Claude’s (often complex and surprising) internal mechanisms.
We are proud to present our new preprint “Correlative light and electron microscopy reveals the fine circuit structure underlying evidence accumulation in larval zebrafish”, just posted on bioRxiv (biorxiv.org/content/10.110…). (1/10)
Can we predict future brain activity in a small vertebrate?🤔 We're releasing ZAPBench⚡️(#ICLR2025 spotlight): a benchmark to forecast activity in a whole vertebrate brain🧠 at single-cell resolution!🐟 70k+ neurons, 3 TB of data & extensive baselines. Connectome is coming! 🧵👇
🧠 In #connectomics, artificial neural networks analyze biological ones, but large-scale benchmarks have been lacking. We introduce NISB - the Neuron Instance Segmentation Benchmark, generated with novel 3D diffusion models! 1/2
If you've seen the @FlyWireNews Fly #Connectome images making the rounds and wondering what all the fuss is about, this video is for you. A #scivis primer that covers some essential #neuroscience, context for this historic moment, and the challenge ahead youtu.be/VPaxhoRgqgU
One the cool possibilities from the #fly #connectome is the ability to run simulations. I've finally got around to experimenting with the "Leaky Integrate and Fire" model by @Philip_Shiu et al. Here are test simulations of 3x Mi1 optic cells, followed by their whole columns
🧵on Japan's underrated contributions to neural nets. Shun-ichi Amari @UTokyo_News_en @riken_en is another one of my heroes. His 1972 paper on associative memory models modeled Hebbian plasticity using an outer product weight matrix.
Wow. Hopfield model and Boltzmann machine.
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Physics to John J. Hopfield and Geoffrey E. Hinton “for foundational discoveries and inventions that enable machine learning with artificial neural networks.”
BREAKING: Congratulations to PNI professor emeritus John Hopfield, who was awarded the 2024 @NobelPrize in Physics, alongside @UofT's Geoffrey Hinton “for foundational discoveries and inventions that enable machine learning with artificial neural networks” pni.princeton.edu/news/2024/pnis…
And thanks to Connectomics @Google for creating Neuroglancer, which was indispensable for visualizing failure modes during AI R&D, and also served as the front end for #FlyWire proofreading. x.com/stardazed0/sta…
Neuroglancer (github.com/google/neurogl…) now supports touch controls. Example view: neuroglancer-demo.appspot.com/#!%7B%22layers…
The fly connectome was impossible to achieve under the traditional scientific funding apparatus alone. We'd all be better off with structural changes that make achievements like this more likely in the future. 1/
Little known facts: @SebastianSeung was an early adopter of ConvNets. He taught ConvNets in his computational neuroscience class at MIT in the late 1990s. In the mid/late-2000s, he started his research program on connectomics and wanted to use ConvNets to analyze the image slices…
Some people have gotten the impression that I hate backprop. Let's be real. I love backprop. The fly connectome was enabled by an AI system containing an arsenal of convolutional nets trained by backprop.
Ah yes, what do computer scientists need to know about cell types? In five words or less?
I believe Dona Fetter did the dissection. And that's right, doing this well is a fine art, micro-precision and high speed combined. This video shows the technique: youtube.com/watch?v=dc9tFp…
Re: 'unsung' heroes @zhihaozheng acquired 80% of the images in the FAFB dataset while in my lab, then did beautiful (IMHO) analysis of PN->KC connectivity therein (cell.com/current-biolog…) and now is a postdoc in the @SebastianSeung and Tank labs doing hippocampus! 🙏
Bridging the gap between connectomics and functional understanding requires computational modeling. Shiu et al’s computational model simulates the activity of the entire Drosophila brain, accurately predicting neuronal responses. nature.com/articles/s4158… youtu.be/bZVoPJumx8Y
Since the '50 largest neurons' #connectome render with @amyneurons @FlyWireNews has made it's way around the world, I'm sharing an animated version. Including: Information flow from the ocelli, 50 largest #neurons, and a LOT of #synapses on one MASSIVE neuron. #neuroscience