
Robin Schmid
@rschmid1789
Postdoc in Computational Mass Spectrometry Chief Architect of mzmine Co-Founder mzio GmbH PhD Analytical Chemistry Java, Python, DevOps, ML, cloud computing
Working with @HeuSteffen on the latest #ionmobility #CompMS #metabolomics methods is a blast - dynamic full-stack development with idea ping pong. Meet him at #asms2024. #proudPI
Interested in imaging, metabolites and lipids, and how to do it with confidence? Check out Steffen's poster #362 today at #ASMS2024 on bringing MS/MS to imaging experiments. #SIMSEF #PASEF #mzmine
Our team has just completed the new release of #mzmine 4.5.0, boosting its performance for feature alignment (~2-3x), spectral library matching (~20x), and molecular networking. On my laptop, I ran 250 DOM samples: 10 s for library matching and 23 s for molecular networking.
New mzmine release. Learn more here: mzio.io/mzmine-news/ bsky.app/profile/mzmine… linkedin.com/posts/mzio_mzm…
Join our free @mzmine_project webinar series together with VMOL. Starting off with mzmine 4 for #chromatography #LCMS #massspectrometry #CompMS #metabolomics. November 5th (next Tuesday) Register with VMOL to receive updates and the zoom link: docs.google.com/forms/d/e/1FAI…

Check out the new version @mzmine_project 4.2.0. Especially for all Bruker #timsTOF users with the 2.5x faster data import. Also give us feedback on the optimized GC-MS workflows - as always start with the #mzwizard #metabolomics #MassSpectrometry github.com/mzmine/mzmine/…
youtu.be/7TZH2lng-MI New #mzmine release out now! Check out the latest features added in version 4.2: • Optimized GC-MS workflow • Faster timsTOF data support • Added Bruker .baf support
Interested in Alzheimer's disease animal models and gut microbiome? Don't forget to check the talk of our own @simonezuffa. He will be presenting during the Neurological Diseases session at 2:40 PM #MetSoc2024 #EMNcommittee
I really enjoyed @LF_Nothias's keynote talk on AI chatbot for education and research in metabolomics. There is one secret he didn't show in the demo; actually you can hear the answers in voices of @LF_Nothias, @boeckerlab, or @rschmid1789 🤣 #MetSoc2024 #Metabolomics2024
How better to spend an extended layover than to connect with the Korean #metabolomics and #MassSpectrometry research community. Thanks @kyobinkang for hosting. @corinnabrungs and I will also have some time to enjoy Korean culture and food. Register now and see you at the workshop
Register now for our @mzmine_project #CompMS workshop hosted by @kyobinkang: July 9th, 10 am, Gemma Hall (Changhak B107), @sookmyung1906 University, Seoul, Korea. @corinnabrungs & @rschmid1789 will introduce non-target #MassSpectrometry data analysis. forms.gle/5P16q6dqeDsMtt…
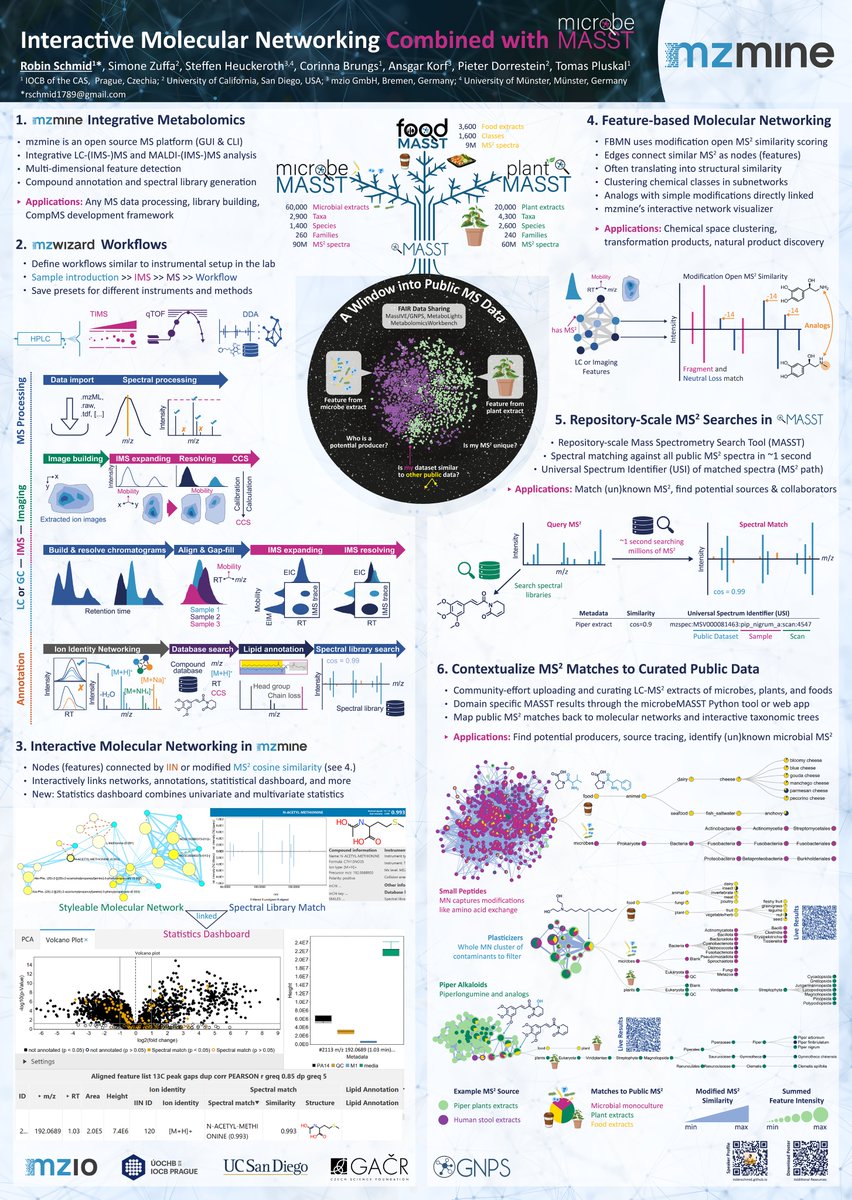
Lunch break is the last chance to visit and discuss my poster #1003 on @mzmine_project and #microbeMASST. Download and more info here: github.com/robinschmid/po…

Find me at my poster #1003 on the interplay of @mzmine_project, @GNPS_UCSD, #microbeMASST, #plantMASST to search (un)known MS2 #MassSpectrometry against all public data. Thanks to @MetabolomicsSoc for the travel award to join #Metabolomics2024 Download: github.com/robinschmid/po…
Join the bioinformatics Hub at #MetSoc2024 by the @MetabolomicsSoc and @EMN_MetSoc I will be there with other people from the mzmine team and collaborators

Register now for our hands-on @mzmine_project workshop at the @MetabolomicsSoc #Metabolomics2024 conference. There are still a few slots left. Meet @cbrungs1789, @rschmid1789, @tomas_pluskal, @damiani_tito, Roman, and @LF_Nothias Just a few steps on: metabolomics2024.org/metabolomics20…
Apply now for an opportunity to work in an international lab with a fantastic PI @tomas_pluskal with ambitious goals in a leading institute @IOCBPrague in one of the greatest cities in the heart of Europe #prague. Two #PostDoc and one #PhD positions.
My lab has several open research positions🪴🧬🔬 Please see the links below and contact me for more information!
Thanks @EMN_MetSoc for highlighting my research! See you all at @MetabolomicsSoc #MetSoc2024 conference in Osaka, Japan. Or during our @mzmine_project workshop at @OISTedu on Okinawa. Reach out to me if you want to join our effort on open mass spectral libraries and #CompMS.
Introducing Robin for #ECRVoices @rschmid1789 is working on #MassSpectrometry and he is based in Prague, Czechia 🇨🇿
My talk on using the #ZenoTOF and #EAD for lipids is finally online. It was recorded last year during #ASMS by @SCIEXnews. sciex.com/applications/b… I bringt it up because, since then, we made great progress on the automated and unbiased annotation of lipids. You can read about…
Introducing Andres for #ECRVoices @a_cumsille is working on #NaturalProducts and he is based in Gainesville, USA 🇺🇸
If sampling the International Space Station sounds interesting to you, join me Monday morning at #ASMS2024 to hear our story!
mzmine protocol just came out! Download the latest mzmine 4, jump into the wizard, and try it on your own #MS data! Special thanks to my best coding pal @HeuSteffen and to @damiani_tito, @tomas_pluskal et al. for the amazing co-working xp on this article. nature.com/articles/s4159…
Check out our protocol on reproducible #MassSpectrometry data processing and compound annotation in @mzmine_project led by @HeuSteffen @damiani_tito @rschmid1789 @tomas_pluskal and bringing together many #OpenSource collaborators. Test it in mzmine 4 now. nature.com/articles/s4159…
