Kang, Kyo Bin
@kyobinkang
Associate Professor at Sookmyung Women's University, Seoul, Korea; A natural product chemist using mass spec to discover chemical diversities in nature.
Today our lab's latest work is published in @PNASNews. We suggest that metabolomics, especially enhanced by @GNPS_UCSD and MassQL, is a powerful method for phenotyping of microbial transformation; but this paper says more. A thread. 1/ pnas.org/doi/10.1073/pn…
🇪🇺🤝🇰🇷 Today, the Republic of Korea joins Horizon Europe. It is the first Asian country to take part. Korean entities can now join and lead research consortia with some of the world’s best research organisations to tackle global challenges together. More info ↓
Finally out! First discussed in 2022, this study shows that metabolites with gut microbial origin are enriched in the blood of PD patients with RBD, a prodromal symptom of Parkinson’s, unlike those with PD only. Happy to be a co-corresponding author. nature.com/articles/s4153…
We discovered that monoterpene indole alkaloids from Vietnamese plant Uncaria scandens inhibit cancer cell motility by suppressing epithelial-mesenchymal transition via integrin α4-mediated signaling. Collaborated with Prof. Hangun Kim. doi.org/10.1038/s41598…

LIVE UPDATE - @MetabolomicsSoc is aware of the traffic situation and has organised bus transport to the conference dinner from Prague Congress Centre #metabolomics2025 #Prague
In this article published today in @metabolomics we introduce the Korea MetAbolomics data rePository (KMAP), and describe what our team has pushed hard and what we will do, to make it standardized and harmonized with the other data repository. doi.org/10.1007/s11306…

Flying to Prague for #Metabolomics2024, the MetabolomeXchange meeting, and some other things. See you soon dear Metabolomics folks!

This paper represents a great effort by @roman_bushuiev and his brother @AntonBushuiev. The DreaMS foundation ML model for mass spectra of small molecules now opens lots of avenues for possible downstream applications. It might be a game changer for computational metabolomics.
Self-supervised learning of molecular representations from millions of tandem mass spectra using DreaMS go.nature.com/4k1n5iC
Big congrats to Raphaël Rodriguez, his lab, and the whole team of collaborators on a fantastic paper on mobilizing lysosomal iron to drive ferroptosis: nature.com/articles/s4158…
I am excited to share the latest project I have been working on "A Multi-Organ Murine Metabolomics Atlas Reveals Molecular Dysregulations in Alzheimer’s Disease". 1/n biorxiv.org/content/10.110…
Really happy to share that our lab's 5-year project on human microbiome metabolites has just been published in @J_A_C_S! pubs.acs.org/doi/10.1021/ja…
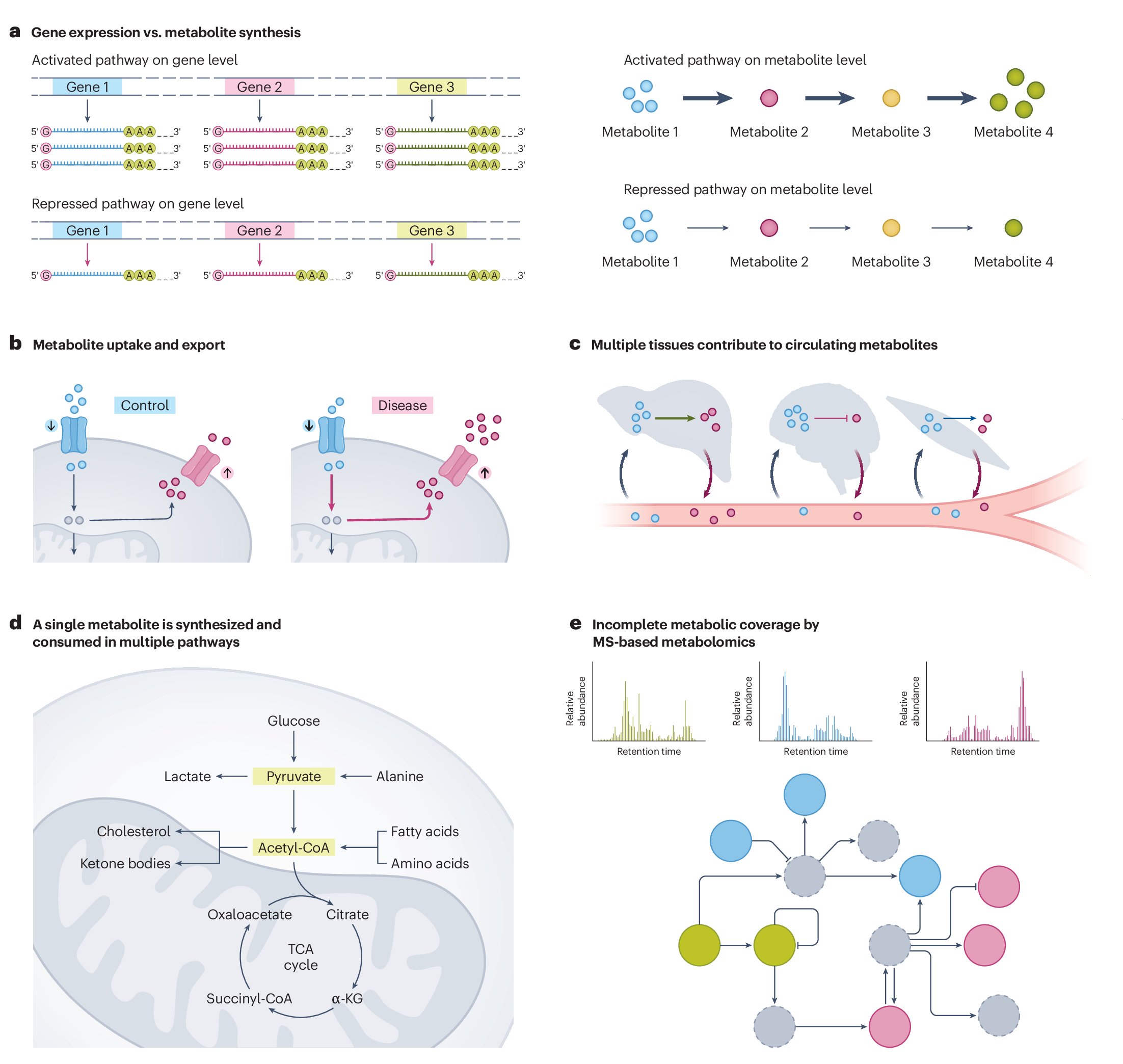
This is a must-read for everyone involved in metabolomics. Personally I love this article because I no longer have to write a long paragraph when I say 'please don't use pathway analysis' during peer review 🤣 nature.com/articles/s4225…
Oliver Fiehn is giving his plenary lecture on his solution for "making metabolomics great again: the LC-BinBase environment" in the Korea Metabolomics Society annual meeting.
