OsmanLab
@osman_lab
Studying the mysteries that surround the genome of the cell's powerhouse
Thrilled to announce that our latest paper on the novel mitochondrial HMG-box protein, Cim1, just got published in @NAR_Open. academic.oup.com/nar/advance-ar… Surprisingly, Cim1 antagonizes the TFAM homolog Abf2 in #mtDNA copy number regulation. Congrats, Simon!! #lmu #mitochondria 🔬

🚀 Excited to share our latest work on Mrx6 and mtDNA copy number regulation, now available on @biorxivpreprint ! We uncover how Mrx6 and Mam33 interact with the Lon protease Pim1 to control substrate degradation. 🔬 Check it out here: biorxiv.org/content/10.110… #mitochondria #mtDNA
Mrx6 binds the Lon protease Pim1 N-terminal domain to confer selective substrate specificity and regulate mtDNA copy number biorxiv.org/content/10.110… #biorxiv_cellbio
🚨 New segmentation model alert! 🚨🥳 You can finally use InstanSeg in our #BioImageAnalysis software #CellACDC! InstanSeg is a fast and multi-channel cell segmentation model by @petebankhead, Thibaut Goldsborough, & co. The multi-channel ability is very interesting!
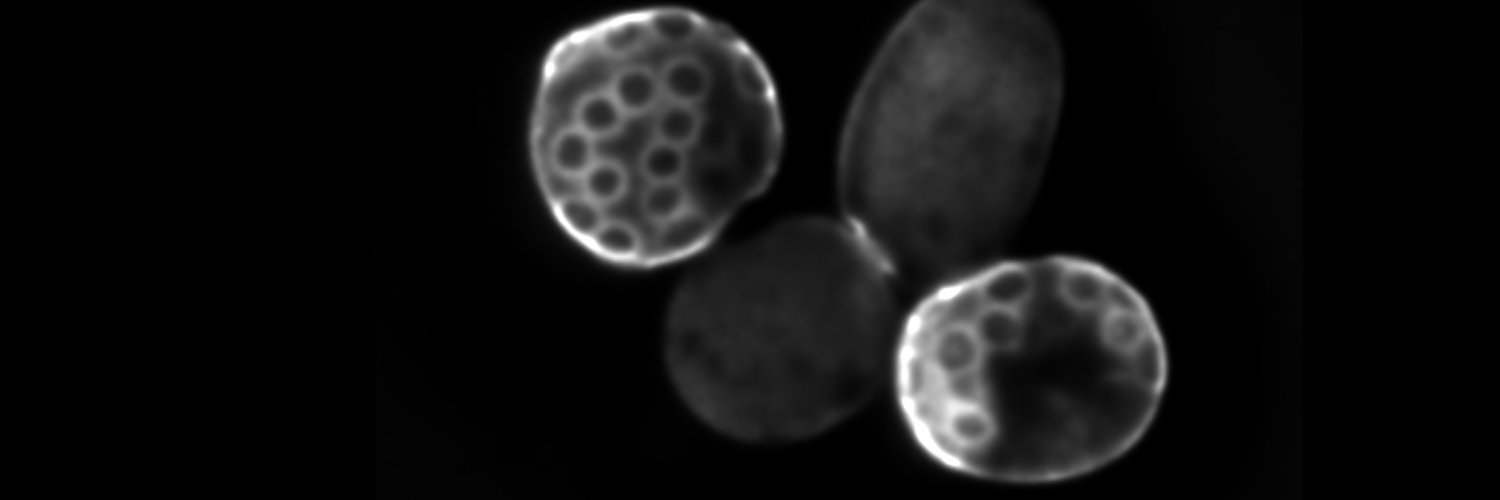
It is not trivial to make the right amount of histones in different nutrients, when #cellsize, growth rate, and S-phase duration change. Still, budding yeast manages by regulating transcription and translation: doi.org/10.1038/s44318… @frank_pado
Huge thanks to @anjbadri and @nitish_dua3 for spotlighting our work in @embojournal ! 🙏🧬 Check out the full article here 🚀🔬 embopress.org/doi/full/10.10… #mtDNA #mitochondria

Disordered regions in the IRE1α ER lumenal domain form condensates in vitro and mediate its stress-induced clustering and signaling in cells Elif Karagöz @gekaragoz @MaxPerutzLabs and coworkers embopress.org/doi/full/10.10…
Glad to share this little piece referring to last paper published in @Nature by Prashar et al. Selective degradation of mitochondrial inner membrane through direct transfer to lysosomes @NatureNV @AndreasReicher8 @ReichertLab nature.com/articles/d4158…
Grateful to everyone who contributed and beyond excited to share my work with you!
Real-time assessment of yeast heteroplasmy dynamics at the single-cell level: Asymmetric partitioning during cell division & limited mitochondrial fusion/fission frequencies drive mtDNA variant segregation @osman_lab @LMU_Muenchen embopress.org/doi/full/10.10…
Introducing a resource of mtDNA copy number (mtCN) across ~10k human donor-tissues (GTEx), 20 murine tissues, and 173 cancer cell lines. Insights emerge by integrating mtCN w/other large-scale measures, e.g., DepMap cells w/high mtCN resist loss of GPX4. pnas.org/doi/10.1073/pn…
Real-time assessment of yeast heteroplasmy dynamics at the single-cell level: Asymmetric partitioning during cell division & limited mitochondrial fusion/fission frequencies drive mtDNA variant segregation @osman_lab @LMU_Muenchen embopress.org/doi/full/10.10…
It was an absolute pleasure to collaborate on this, fantastic work lead by @RoussouRo. I'm very happy to see that our #BioimageAnalysis software #CellACDC was useful to analyse these fantastic images @SchmollerLab
🚀Excited to share our latest paper, where we explore dynamic changes in mtDNA heteroplasmy in living cells! Discover how asymmetric partitioning and mitochondrial dynamics drive mtDNA variant segregation! 🔬embopress.org/doi/full/10.10… #Mitochondria #microscopy #microfluidics #mtDNA
🚀Excited to share our latest paper, where we explore dynamic changes in mtDNA heteroplasmy in living cells! Discover how asymmetric partitioning and mitochondrial dynamics drive mtDNA variant segregation! 🔬embopress.org/doi/full/10.10… #Mitochondria #microscopy #microfluidics #mtDNA
Just wrapped up an amazing retreat at Schloss Ringberg🤩! With fantastic guest speakers/lab alumni, we enjoyed discussing all our exciting projects, hiking, and playing games in this cosy place🍻☀️! #LabLife #scienceretreat #mitochondria #researchlife #science
Just back from a wonderful retreat with the @osman_lab in Tjärnö on the Swedish west coast @GothenburgMBL. Three days of scientific discussions around mitochondrial gene expression and maintenance, beautiful scenery and lots of fun. Thanks to everybody for the good times!!
Awesome fun activity at the retreat with the @Ott_MitoLab at Tjärnö Marine Station 🔬🪸🪼🦀🦭

mtDNA heteroplasmy dynamics are so complex, even paradoxical. To understand why, we combined precise mtDNA editing w/a new method SCI-LITE. In dividing cells, selection (not drift) acts on cell (not intracellular) fitness to shape population heteroplasmy. rdcu.be/dFDCY
Excited to share major progress on next generation of AlphaFold! Together with @IsomorphicLabs, we’ve significantly advanced accuracy & expanded coverage beyond proteins to other key biomolecular classes. Continuing to accelerate science at digital speed dpmd.ai/x-alphafold-pr…
Happy Halloween everyone!!! Thanks @DvaliTamara for organizing!!! Traditional pumpkin carving together with @MokranjacLab and Böttger labs! 🎃
