Ophir Shalem
@ophirshalem
Scientist, Associate Professor, Department of Genetics, University of Pennsylvania, Children's Hospital of Philadelphia
Check out our paper, now extensively revised and published in @MolecularCell! This has been a real tour de force led by @stephsansbury and @YSereb and we are very excited about the findings and future potential of this approach cell.com/molecular-cell…

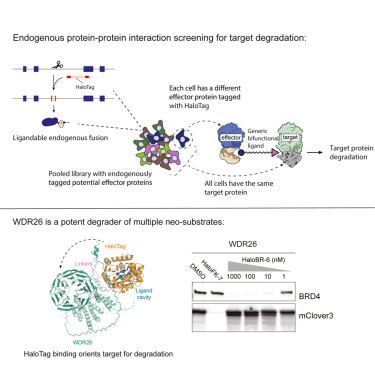
Excited to share this collaboration spearheaded by @GMBurslem lab! Part of our long term goal of developing tools for scalable and non-disruptive perturbation and imaging of the endogenous proteome!
Ever wanted to edit a protein sequence inside a living mammalian cell? Maybe to site specifically incorporate a non-canonical residue in a cell? Perhaps you’d like to do a pull down without an antibody? Or use a small molecule fluorophore to image an endogenous protein?
New in @ScienceMagazine: Our paper mapping gene/isoform/splicing regulation at pop-scale during human #neurodevelopment. Major developmental shifts in #genetic reg., New colocalized risk genes at ~60% of #psych #GWAS loci. #PsychENCODE24 science.org/doi/10.1126/sc…
Scouring the human Hsp70 network uncovers diverse chaperone safeguards buffering TDP-43 toxicity biorxiv.org/content/10.110… #biorxiv_biochem
Check out our work on protein editing!
A pair of split inteins—small protein segments that can undergo a self-sustained protein splicing reaction—enables endogenous protein editing in living mammalian cells, according to a new study in Science. 📄: scim.ag/3ENypj9 #SciencePerspective: scim.ag/430jwC9
Excited to to see our collaboration with @GMBurslem lab published today! This is part of our ongoing long term goal of achieving scalable non-disruptive and direct imaging and perturbation of the endogenous proteome science.org/doi/10.1126/sc…
Super excited to launch nimbusimage.com, our cloud-based image analysis platform! nimbusimage.com Docs here: docs.nimbusimage.com Free tier available!
Huge congratulations to former lab postdoc @YSereb for starting his own independent lab! Make sure to follow him for exciting future science and check out his lab if you are looking for opportunities in the NJ/NY area!
The Serebrenik Lab is hiring! Join us in unraveling proteostasis with pooled protein tagging and manipulation! Interested postdocs can learn more & meet other excellent labs at the Rutgers Postdoc Peek (Apr 3-4, 2025). Apply by 1/8/25: rutgers.ca1.qualtrics.com/jfe/form/SV_en…
Fascinating work from Jongens and @ErikaHolzbaur lab demonstrating that FMRP granules mark mitochondrial fission sites in axons and dendrites and serve as sites of local translation!
Incredibly proud of the beautiful work from Adam Fenton!
🚨🚨🚨 Thrilled to share our newest study — identifying functional long noncoding RNAs using transcriptome-scale RNA-targeting CRISPR screens. 🔎🔎🔎 We hope this scalable approach will be helpful for folks who want to connect noncoding RNAs to phenotypes & diseases.
Now online! Transcriptome-scale RNA-targeting CRISPR screens reveal essential lncRNAs in human cells dlvr.it/TG6t4q
Excited to see the revised version of our paper now published at @CellGenomics led by @YSereb cell.com/cell-genomics/…
Surprise - FTLD-TDP Type C is a dual proteinopathy! Annexin A11 co-accumulates with TDP-43 in all cases of FTLD-TDP Type C. #annexinopathy #FTD #dementia #ADRD #ActaNeuropathologica doi.org/10.1007/s00401…
Such a fun evening with bittersweet goodbyes and welcoming new members. Real privilege to work with so many talented scientists. Make sure to follow @stephsansbury @YSereb @Saranya to find out about the next steps in their careers!
