
Thomas Kislinger
@KislingerThomas
Clinical Proteomics, Cancer Biology & Mass Spectrometry @MBPatUofT and @pmcancercentre 🇨🇦
checkout our latest paper in @NatureBiotech. We developed a graph-based algorithm that comprehensively generates non-canonical peptides in linear time. nature.com/articles/s4158…

Proud to share our new @Nature study: We developed the 1st NNMT inhibitor that reprograms cancer-associated fibroblasts, boosting immune response & stopping tumor growth in ovarian cancer models, especially when paired with immunotherapy. @OvCa_UChicago nature.com/articles/s4158…
Our paper on moPepGen is finally out in Nature Biotechnology! moPepGen is a fast graph-based tool for systematically predicting and detecting non-canonical peptides, revealing the “dark side” of the proteome. Hoping it opens new doors in proteogenomics. nature.com/articles/s4158…
New collaborative work from PhD student Aastha. Using proteomics for CSF-based biomarker discovery @DrAMansouri Cerebrospinal fluid protein biomarkers are associated with response to multiagent intraventricular chemotherapy in … pubmed.ncbi.nlm.nih.gov/40321620/

Read our latest #preprint on @researchsquare: Systematic evaluation of analytical methods for CSF proteomics researchsquare.com/article/rs-703…
Phenotypic screens for SIRPA expression reveal RAB21 as a general regulator of macrophage surface identity: Cell Reports cell.com/cell-reports/f…
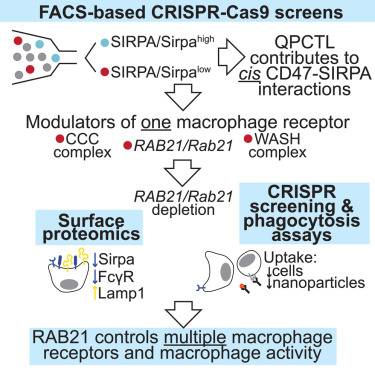
Phenotypic screens for SIRPA expression reveal RAB21 as a general regulator of macrophage surface identity dlvr.it/TLfG4z
Just came across a fantastic article covering our work on moPepGen — truly honored by the thoughtful writing and high praise. Grateful to see our efforts in advancing proteogenomics recognized like this. forwardpathway.us/mopepgen-a-new…
Proteogenomics algorithm improves the detection of hidden genetic mutations #NBTIntheNews via @GENbio genengnews.com/topics/artific…
Identification of non-canonical peptides with moPepGen pubmed.ncbi.nlm.nih.gov/40523945/

A new tool, moPepGen, developed by @uhn @pmcancercentre & @UCLAHealth, can predict & detect unusual proteins involved in cancer and beyond, deducing what cellular processes might have gone wrong to give rise to these mutant peptides. @NatureBiotech: doi.org/10.1038/s41587…
checkout our latest paper in @NatureBiotech. We developed a graph-based algorithm that comprehensively generates non-canonical peptides in linear time. nature.com/articles/s4158…
Identification of non-canonical peptides with moPepGen go.nature.com/3HYJyyz
We are proud to announce the publication of CellNEST, deep-learning detection single cell-cell or even "relay" communication on #SpatialTrascriptomics data at single-spot or #SingleCell! Paper: nature.com/articles/s4159… Code: github.com/schwartzlab-me… and github.com/schwartzlab-me…
Paper(s) alert from our @RESOLUTE_IMI project – In a compilation of four papers in Molecular Systems Biology we show how we were deciphering cellular logistics and present a comprehensive systematic blueprint of chemical transport pathways with #SoluteCarrier transporters
bit.ly/4lSlL2S CNS lymphoma is a challenging disease to manage. Intraventricular chemotherapy may hold promise, but not all patients are candidates. Using #CSF #proteomics, here we show that 1) LCP1 may be candidate predictive marker of those who will benefit and 2)…
New review from our team Recent implementations of data-independent acquisition for cancer biomarker discovery in biological fluids pubmed.ncbi.nlm.nih.gov/40227112/

Today at #AACRprecmed25, Thomas Kislinger will speak in Plenary Session 6: New Omics for Precision Medicine. Read his work in Cancer Research Communications: brnw.ch/21wRiFy @kislingerthomas #prostatecancer #radioresistance #proteogenomics #genomics #openaccess
Bernhard Küster, Thomas Kislinger, and M. Celeste Simon discussed "New Omics for Precision Medicine" in a plenary session at the AACR Special Conference on Functional and Genomic Precision Medicine in Cancer. #AACRprecmed25 @KislingerThomas