
Qian Cheng
@sgqcheng
Senior Editor @NatureComms. PhD @Cambridge_Uni. Views my own.
The @NatureConf on AI-Augmented Biology is open for registration! We will explore how artificial intelligence is transforming the life sciences. 📅 October 22-24 2025 | 📍 Nanjing, China @NJU1902 @Nature @NatureComms @NatureBiotech @naturemethods natureconferences.streamgo.live/ai-augmented-b…

CausCell: a causal disentanglement framework that generates explainable, generalisable, and controllable representations for #singlecell omics data. @greysonc98 @Kejing_Dong #BiotechNatureComms doi.org/10.1038/s41467…

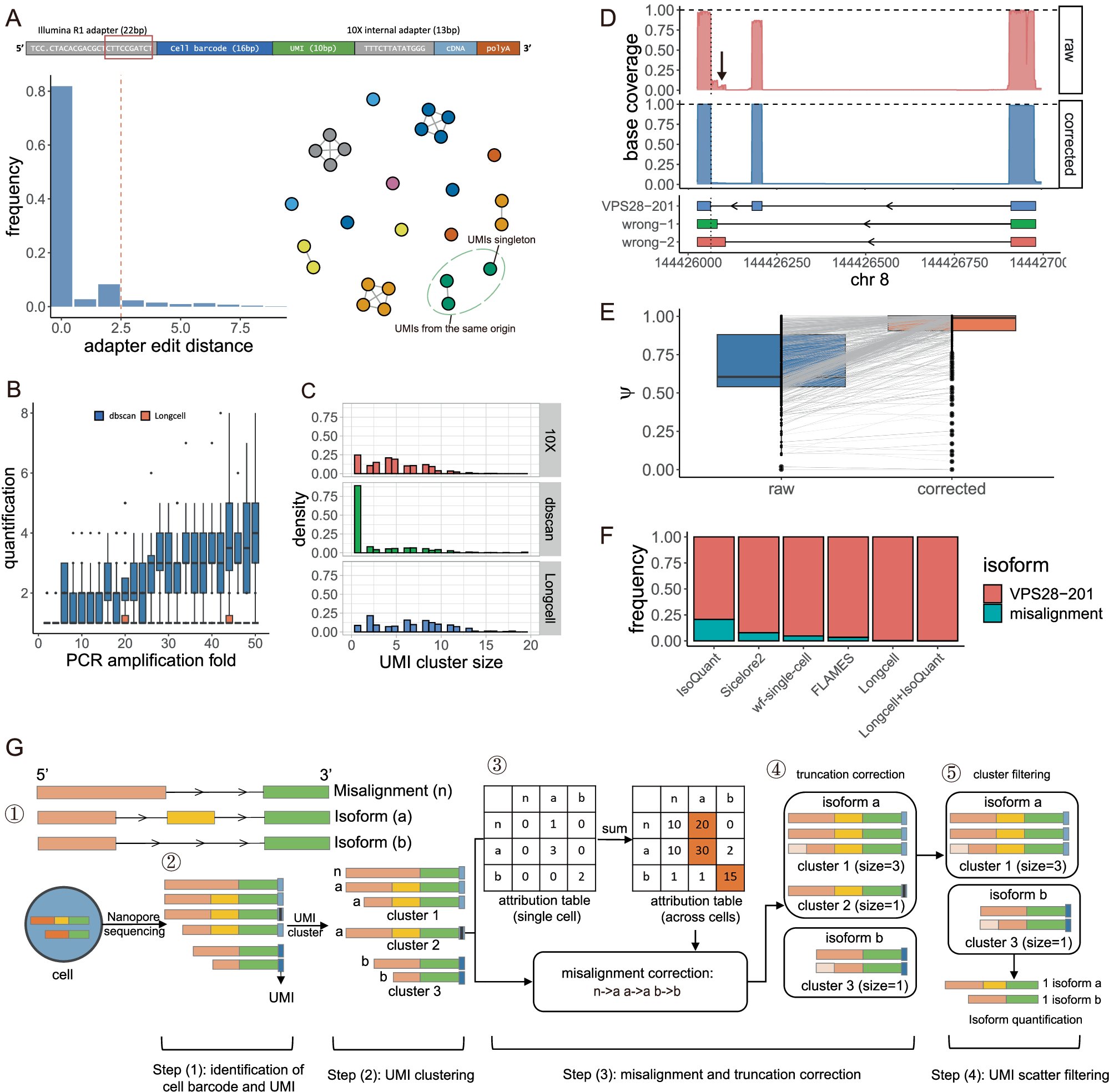
A computational framework for single cell and spatial alternative splicing analysis with Nanopore long read sequencing @NancyZh60672287 #BiotechNatureComms nature.com/articles/s4146…
Bering performs joint cell segmentation and annotation for spatial transcriptomics with transferred graph embeddings @Kang_Jin__ @JianShuLab #BiotechNatureComms nature.com/articles/s4146…

A computational tool #SCALPEL for #isoform quantification at the single-cell level using 3’ scRNA-seq data @miriplass @PlassLab #BiotechNatureComms doi.org/10.1038/s41467…

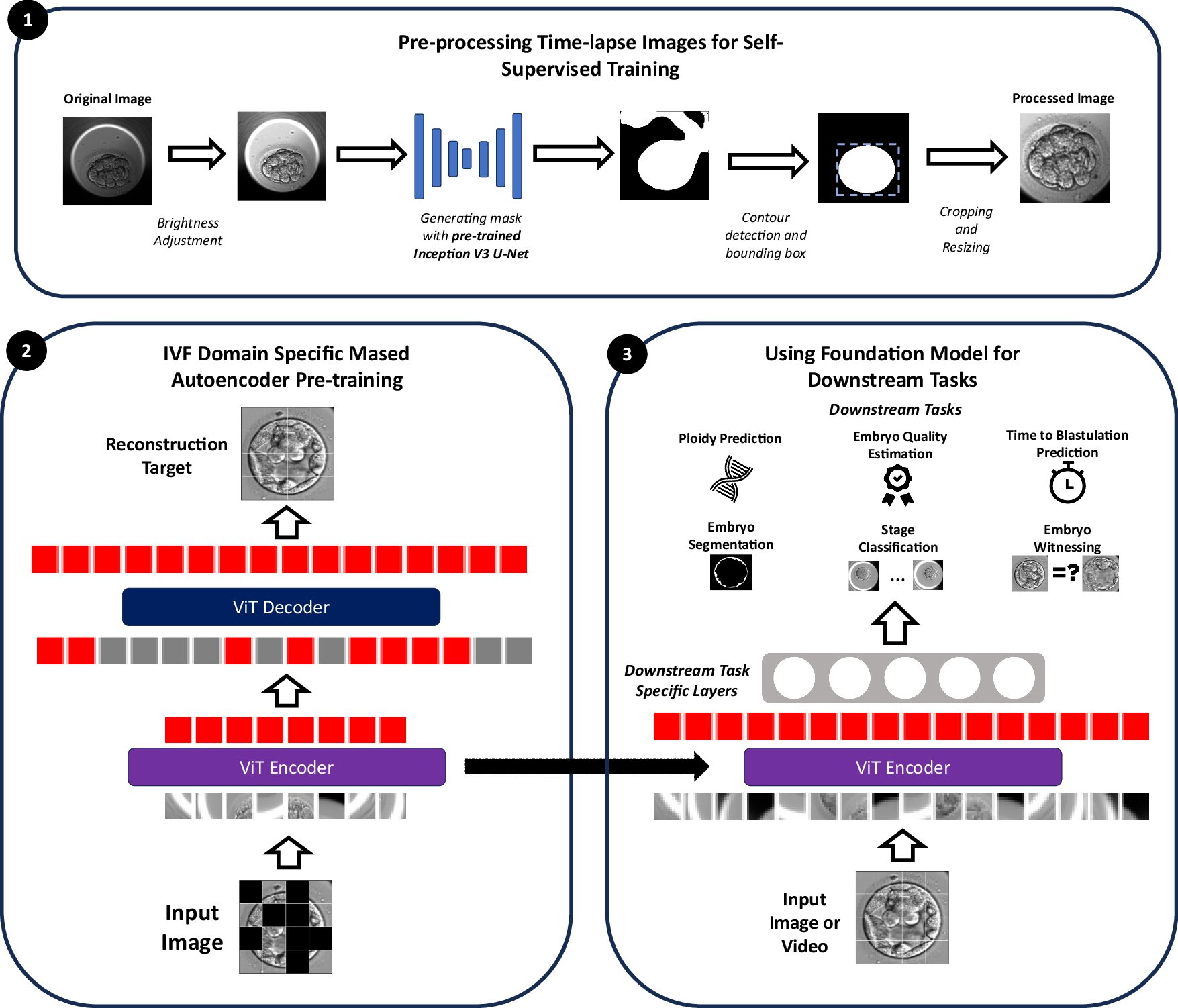
A foundation model trained on 18 million time-lapse embryo images for in vitro fertilisation @rajsuraj99 @hajirasouliha #BiotechNatureComms doi.org/10.1038/s41467…
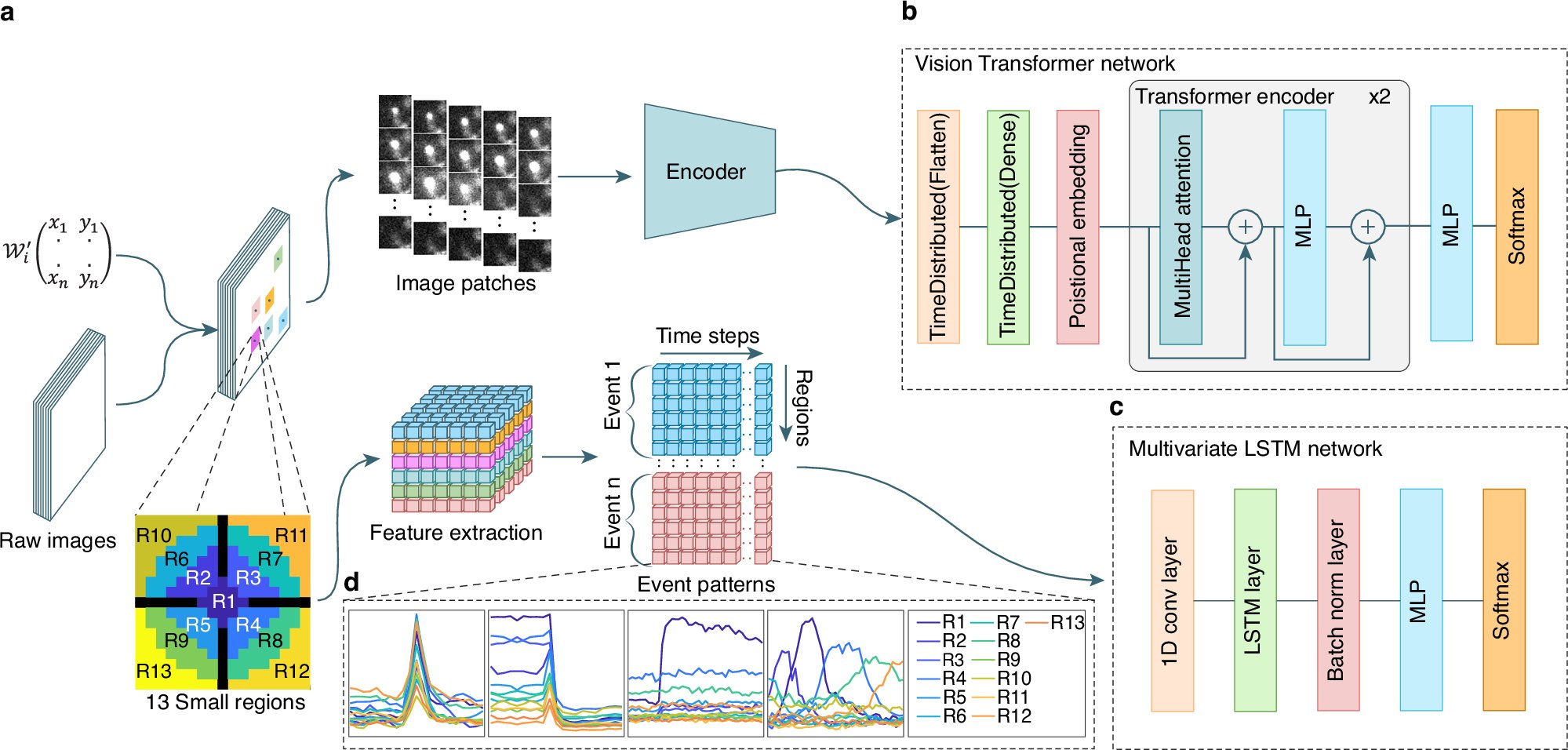
A fully automated AI ImageJ plugin that detects and classifies #exocytosis events from synaptic transmission to single-vesicle fusion. @AliHShaib @AbedShaib #BiotechNatureComms doi.org/10.1038/s41467…
Great to deliver the Nature MasterClasses with Jie @JPanPanJ for the School of Computer Science at Northwestern Polytechnical University in Xi’an, to share our views on what makes a strong computational paper for @NaturePortfolio journals. #labvisit #openscience




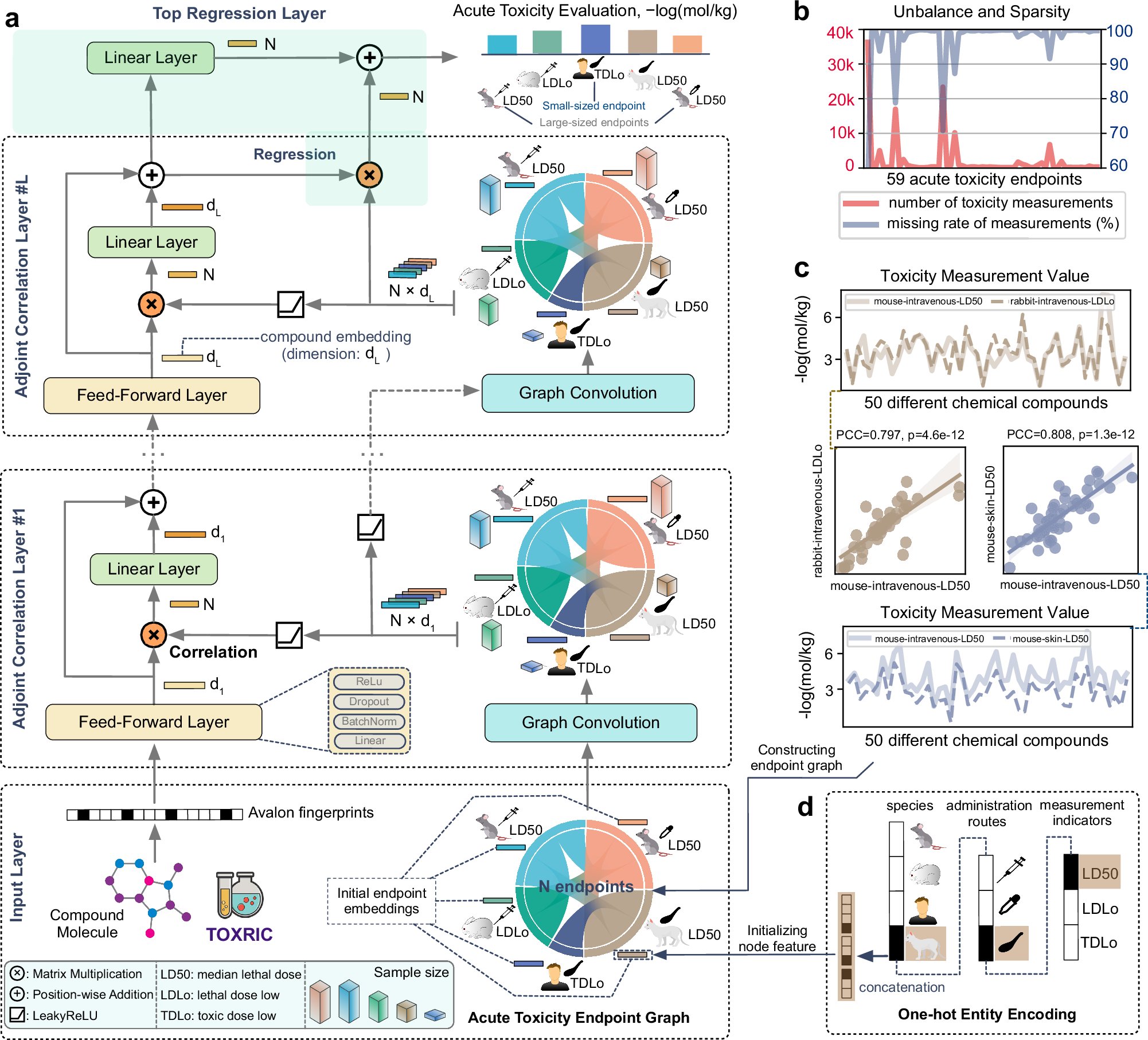
ToxACoL is an endpoint-aware and task-focused compound representation learning paradigm for #Acute_Toxicity_Assessment #BiotechNatureComms doi.org/10.1038/s41467…
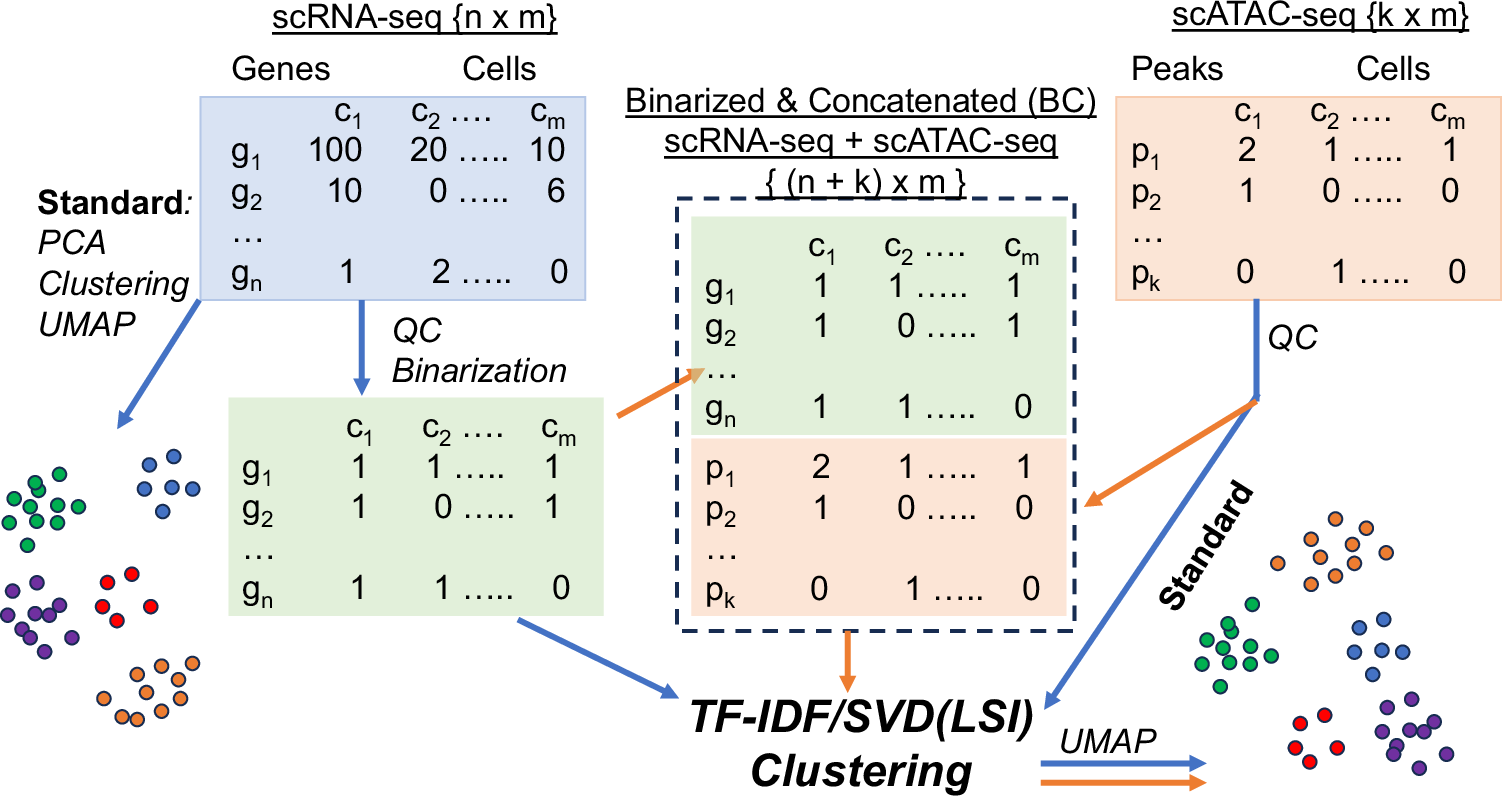
Binarized gene expression data can be concatenated with chromatin accessibility data for effective and integrated cell clustering @zhengdy @rohanmisra @EinsteinMed #BiotechNatureComms doi.org/10.1038/s41467…
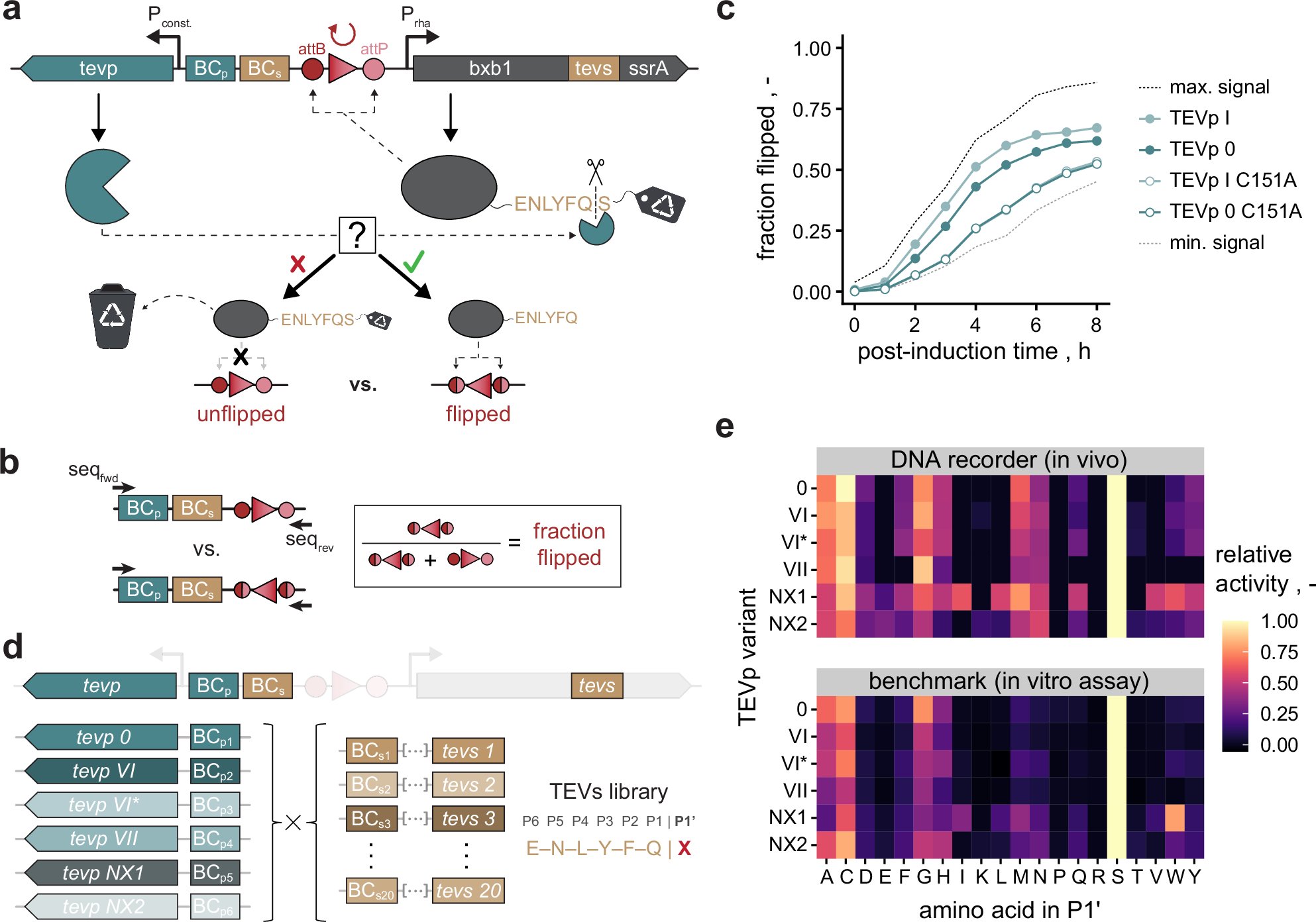
.@JeschekLab describe a data-driven approach combining DNA recording and epistasis-aware deep learning to augment #protease specificity engineering at large scale #BiotechNatureComms doi.org/10.1038/s41467…
A DNATag-based system for secure traceability, featuring error tolerance, mobile phone readability, and robust forgery protection. @jeffnivala @yuan0 #BiotechNatureComms doi.org/10.1038/s41467…

GAUDI integrates diverse omics data types and identifies biologically meaningful patterns across multiple benchmarks. @polcastellano_ @matthewhirschey #BiotechNatureComms doi.org/10.1038/s41467…

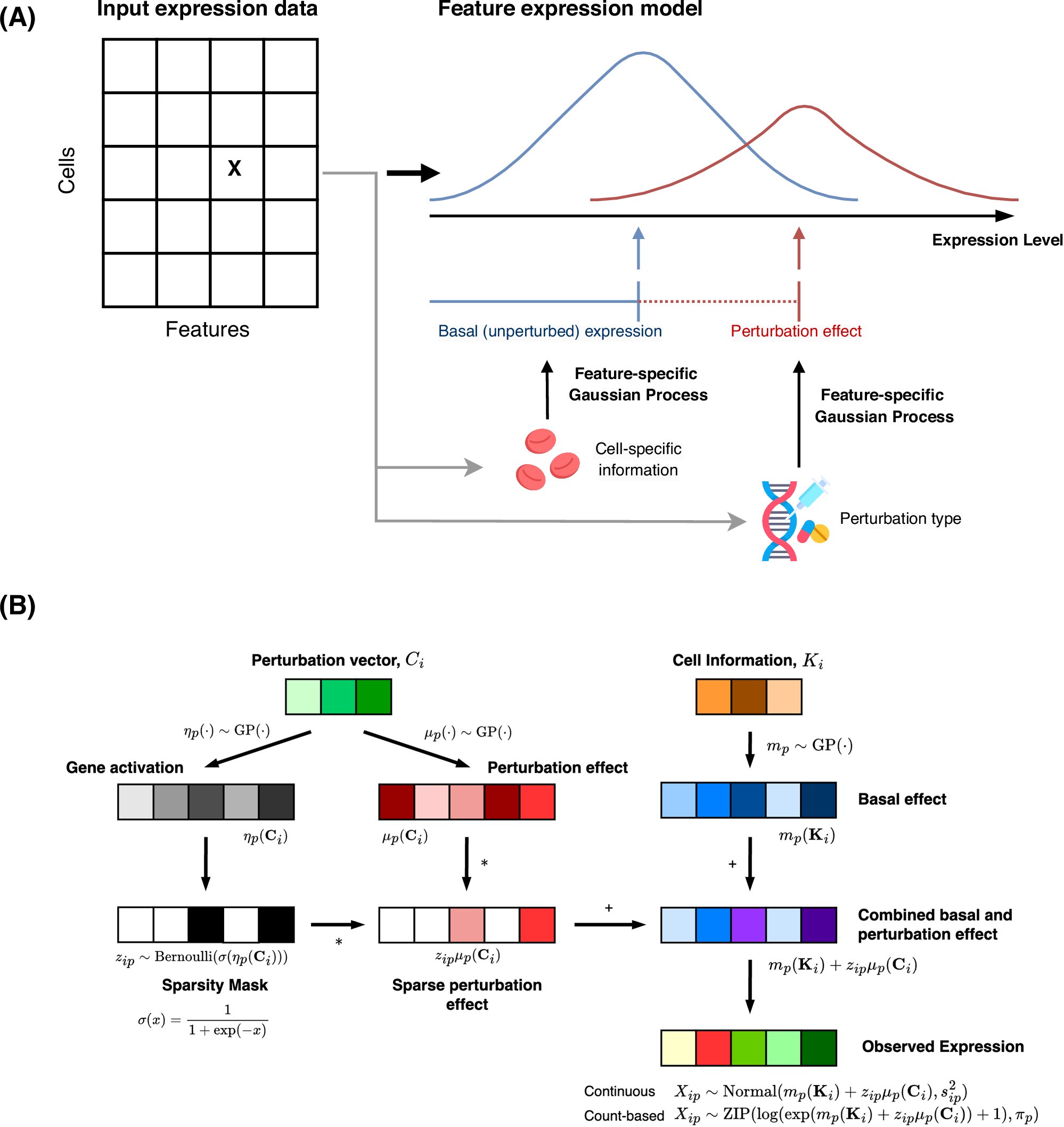
GPerturb detects and quantifies gene-level effects of perturbations, with uncertainty estimates, revealing complex biological interactions. @cwcyau #BiotechNatureComms doi.org/10.1038/s41467…
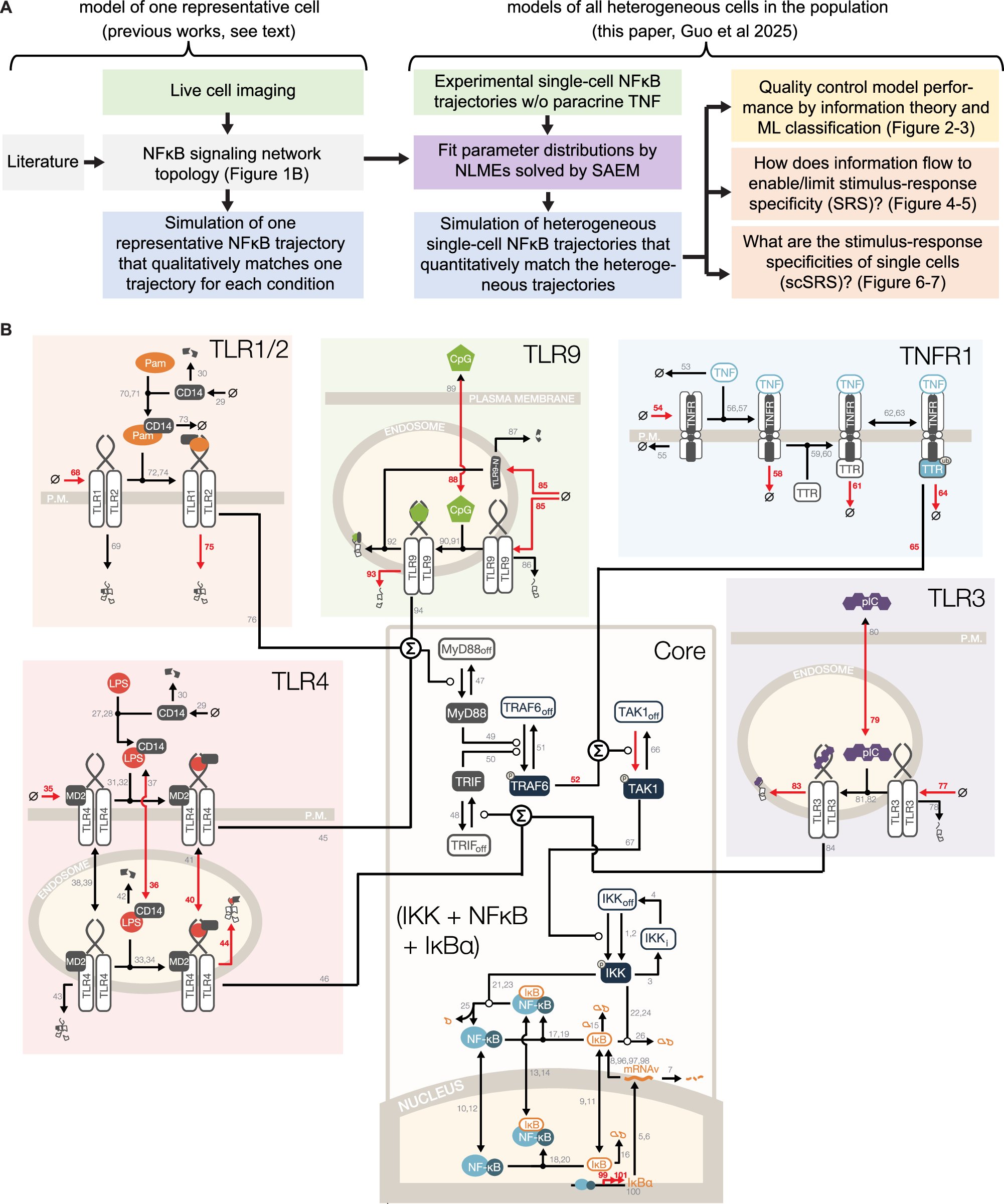
A virtual single-cell NFκB signalling networks that recapitulate heterogenous experimental stimulus-response dynamics #BiotechNatureComms doi.org/10.1038/s41467…
.@Yuxi_Ke @WJGreenleaf develop a high-throughput DNA melting method and improve computational models for predicting DNA folding energies from sequence data #BiotechNatureComms doi.org/10.1038/s41467…

scICE enhances clustering reliability and efficiency for scRNA-seq data analysis @gimhyeo2951042 @umichkim @ibsbimag @IBS_media #Bioinformatics #BiotechNatureComms doi.org/10.1038/s41467…

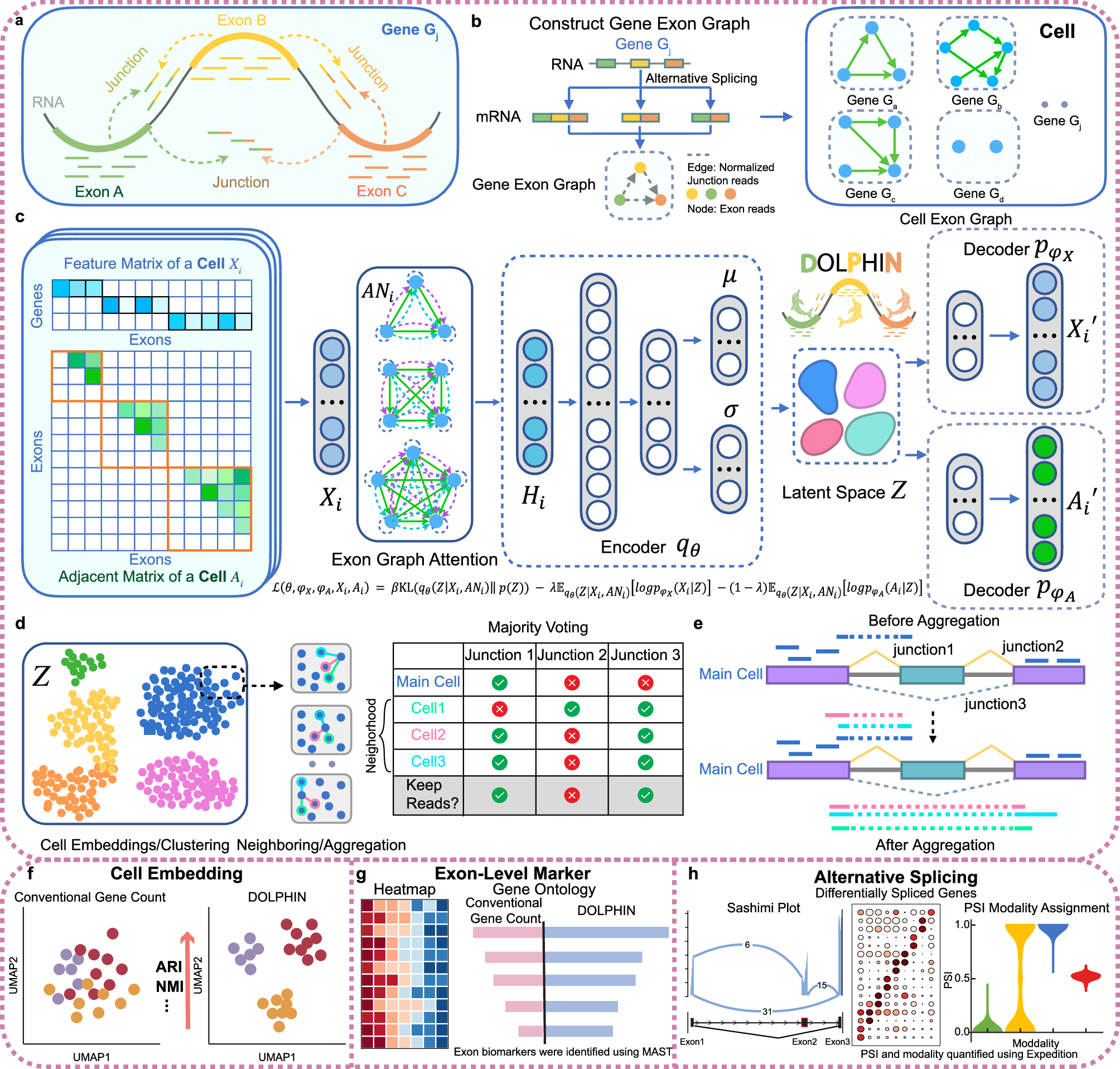
DOLPHIN: a deep learning method that enables exon- and junction-level analysis to improve cell representation and detect alternative #splicing. @Srrrrrrrrrrrr @johnding86 #BiotechNatureComms doi.org/10.1038/s41467…
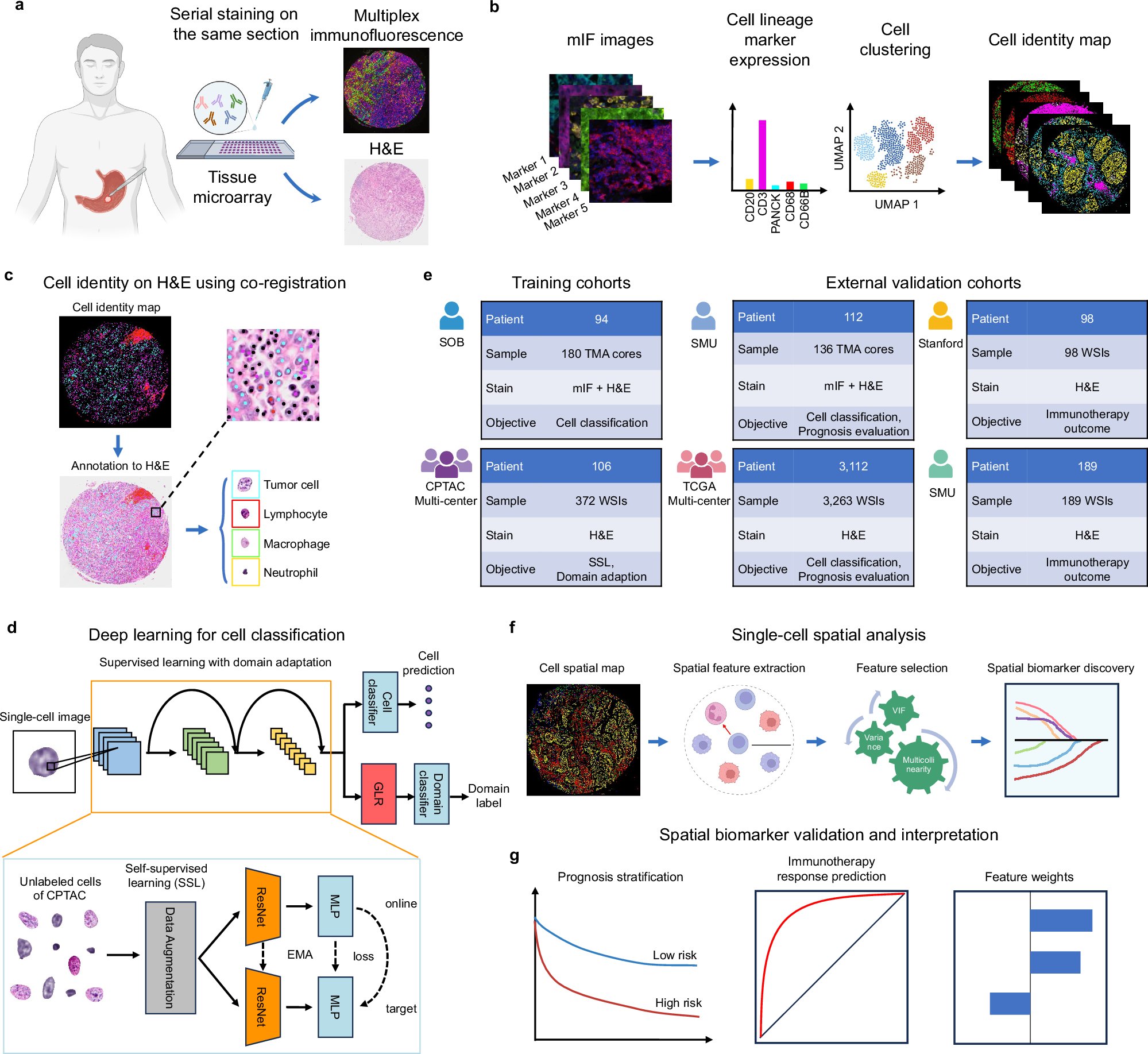
A deep learning and immune-fluorescence-guided cell labelling method for accurate cell pattern identification. #BiotechNatureComms doi.org/10.1038/s41467…
Cell Marker Accordion is a user-friendly platform providing robust automatic cell annotation of #single_cell and spatial populations @RDDS_Lab #BiotechNatureComms doi.org/10.1038/s41467…
