Joseph Replogle
@josephmreplogle
MD PhD, PGY1 at @MGHMedicine/@HarvardMed
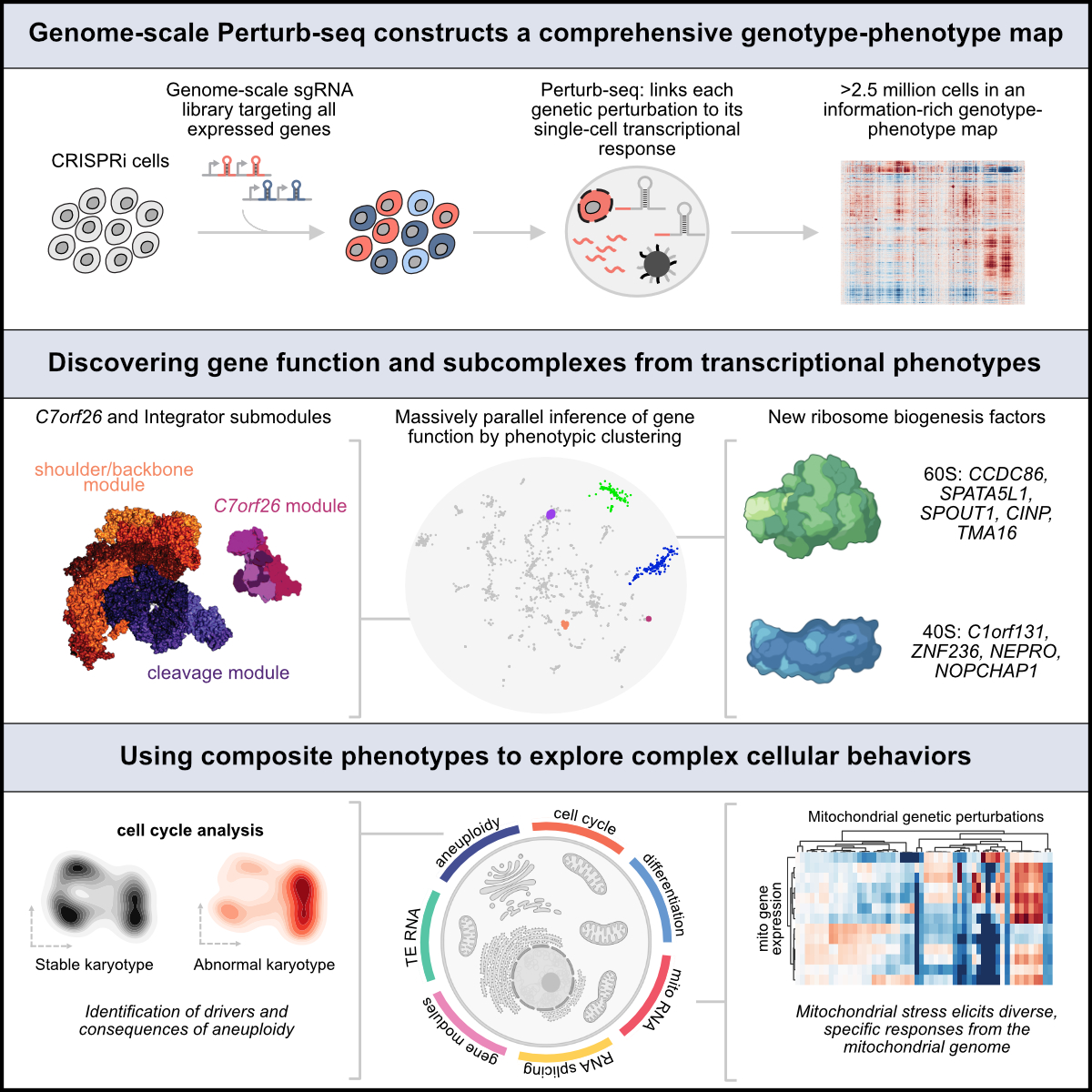
Our genome-scale Perturb-seq paper is out today in Cell! cell.com/cell/fulltext/… @CellCellPress @JswLab Thanks to all the authors and reviewers for their contributions. See the thread below for key findings.
Congrats @NadigAjay on TRADE out now in @NatureGenet: nature.com/articles/s4158… These statistical metrics enable more meaningful comparisons in Perturb-seq atlases. Also, now find the HepG2 and Jurkat Perturb-seq datasets on GEO GSE264667!

Even while in residency and with infants at home, I find it hard not to put everything down to read this new analysis of our data by @jkpritch and team! And in the same week others are working toward optimizing large-scale Perturb-seq in cardiomyocytes biorxiv.org/content/10.110…
Modern GWAS can identify 1000s of hits but it can be hard to turn this into biological insight. What key cellular functions link genetic variation to disease? I'm excited to present our new work combining associations + Perturb-seq to build interpretable causal graphs! A🧵
The Norman lab's new large-scale CRISPRa Perturb-seq is a beautiful combination of question driven biology with single cell tech/analytic developments. I would expect nothing less from the lab!
Can we recreate the transcriptional states seen in single-cell atlas projects? In our new preprint we try to do this using large-scale Perturb-seq experiments. Along the way we discuss off-target effects of CRISPR epigenetic editors and identify drivers of key fibroblast states.
Can we recreate the transcriptional states seen in single-cell atlas projects? In our new preprint we try to do this using large-scale Perturb-seq experiments. Along the way we discuss off-target effects of CRISPR epigenetic editors and identify drivers of key fibroblast states.
Excited to share our preprint "Multiome Perturb-seq unlocks scalable discovery of integrated perturbation effects on the transcriptome & epigenome". This work extends single-cell CRISPR screens to simultaneously profile gene expression & chromatin accessibility.
Just how impactful are large-scale perturbational data in single cell genomics? In the last two weeks, there have been *three* modeling/stats preprints with qualitatively different questions and insights for the same dataset generated by @josephmreplogle @JswLab (links below)
How do genetic perturbations change cells? How are these effects shaped by cell type and dosage? How do we best extract insight from modern massive perturbation atlases? Im pleased to share a new preprint where we develop a suite of statistical approaches to these Qs (link below)
Check out our latest large-scale Perturb-seq on biorxiv! biorxiv.org/content/10.110… A new stat gen/GWAS-inspired analytic framework + large-scale screens in an expanded range of cell types Thanks @NadigAjay and @Luke0connor for involving us!

From @russelltwalton @BlaineyLab , a very useful vector for those interested in taking advantage of sgRNA multiplexing for single cell CRISPR screens!
Our new preprint describes “CROPseq-multi”: a versatile solution for multiplexed perturbation and decoding in pooled CRISPR screens doi.org/10.1101/2024.0… Reagents on Addgene: addgene.org/browse/article…
Today, @NEJM published our clinical study of a novel #CARTCell in patients with #Glioblastoma. Early results with CARv3-TEAM-E suggest safety and bioactivity. Special thanks to all collaborators @MGHNeurosurg and @MGHCancerCenter! nej.md/3VnjUIq
Our study is out @Nature. We identify non-lipid risk pathways for coronary artery disease using pooled CRISPR-interference for 2,285 genes at GWAS loci. With an amazing team we identified the genetic risk pathways for CAD that act in endothelial cells. nature.com/articles/s4158…
Our latest CRISPRi screening tools + tips w/ @JSWlab and @MarcoJost_: (1) dual-sgRNA libraries ⬆ knockdown while decreasing library size (2) comparison of CRISPRi effectors finds that Zim3-dCas9 offers ⬆ knockdown with minimal toxicity (3) validated Zim3-dCas9 cell lines
Maximizing CRISPRi efficacy and accessibility with dual-sgRNA libraries and optimal effectors biorxiv.org/cgi/content/sh… #bioRxiv
Dreaming of a day when you won't need pipette arm PT after doing a genome-scale Perturb-seq experiment 😆
New preprint out! We present a rapid and highly scalable scRNA-seq method. Thousands of samples or millions of cells with only a vortexer. Microfluidics-free single-cell genomics with templated emulsification biorxiv.org/content/10.110…
Fun to see the Perturb-seq toolbox continue to expand from @satijalab @nevillesanjana. Cas13 Perturb-seq has been on the tech with list! Capture of barcode gRNA at 3' end of tandem Cas13 guide array = less sgrna recombination and higher lenti titers.
We are excited to introduce CaRPool-seq, a new technology that utilizes Cas13 to perform combinatorial single-cell perturbation screens! Check it out, esp. if you're interested in simultaneously perturbing multiple genes and exploring genetic interactions: biorxiv.org/content/10.110…