Jesse Engreitz
@jengreitz
Assistant Professor @stanford Genetics and BASE Initiative. Mapping the regulatory code of the human genome to understand heart development and disease.
Our latest in @Nature: Convergence of coronary artery disease genes on endothelial cell programs nature.com/articles/s4158… Incredible work led by Gavin Schnitzler and @kanghelenyihua — a long-standing partnership between our lab and @Dr_RajatGupta My comments below: 1/

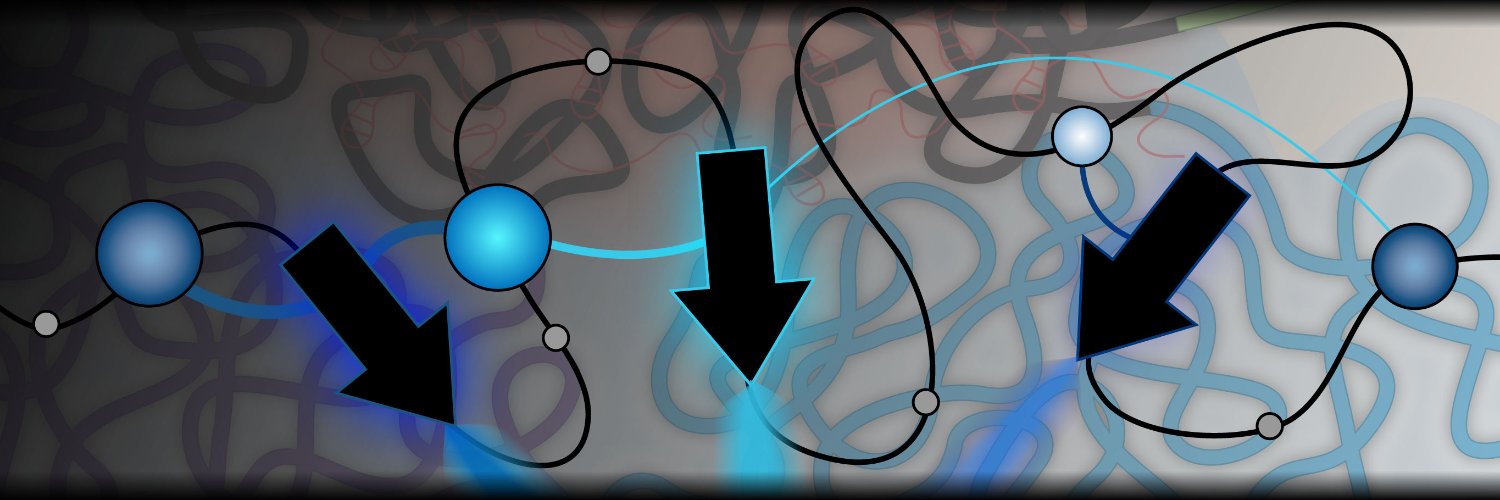
Our latest preprint: Cohesin-mediated 3D contacts tune enhancer-promoter regulation We put the looping model of enhancer function to the test via enhancer CRISPR screens in a cohesin degron cell line See here 👇
I am so excited to share this part of my PhD where we tackle the question: What impact do 3D contacts have on how enhancers regulate their target genes? biorxiv.org/content/10.110… 1/
Here are 10 ways that Trump’s Big Ugly Bill will make your life more expensive: 1. Your utility bills go up as we lose clean energy investments and electricity costs more; 2. Your rent goes up as predatory housing companies artificially jack up prices;
1. What you see here is clearest illustration why things are so messed up and why it cannot get any better until this asymmetry is addressed. Put simply: right-wing has narrative dominance. My org (@mmfa) did this study and been sounding this alarm for years. Some thoughts...
Giving infectious disease research a break, as promised by the MAHA agenda. Let's break down what exactly this Executive Order is *really* saying. Fortunately, I speak fluent anti-vax grifterese & can translate. whitehouse.gov/presidential-a…
Donald Trump is making a deliberate attempt to overwhelm the system, and to overwhelm you. It can be difficult to keep up. And that’s by design. But we can keep up – and respond. Here’s what he’s doing, and what it means for you. Just the facts. 🧵⤵️
Introducing scE2G: a new model to link enhancers to target genes using single-cell data. Excited that scE2G will enable building enhancer maps in hundreds of cell types in the human body! Wonderful collaboration with @robin_andersson @613weilin @mayayayas and others 👇
Excited to share our latest preprint on scE2G – a new model to link enhancers to target genes using single-cell data – with state-of-the-art performance across multiple perturbation benchmarks. biorxiv.org/cgi/content/sh… Read more below! 1/12
Happy to share our new work describing DESynR genes! The evolution of new human protein-coding genes relies on repurposing protein domains from ancestral genes, rather than de novo sequence creation. New genes evolved from old parts, and while we have methods that enable…
1/14 Excited to introduce PertEval-scFM! It provides benchmark and evaluation tools for perturbation effect prediction models, including single-cell foundation models (scFMs). Paper and GitHub link at the end of the thread! 🧵👇
Today, I'm excited to present the second big chunk of my Ph.D. work. We are building a thorough assessment of the burgeoning field of gene expression forecasting. Something is deeply wrong.🧵 - Preprint: biorxiv.org/content/10.110… - Code: github.com/ekernf01/pertu…
Air purifiers in Finnish daycare reduced children's illnesses by 18% in a carefully controlled study. This implies huge potential for better public health and cost savings from sick leaves, with a simple intervention against airborne infections. yle.fi/a/74-20089670
Excited to share the white paper for the IGVF Consortium, supported by @genome_gov Deciphering the impact of genomic variation on function rdcu.be/dSZdm
Understanding the function of an entire genome is no easy task! Most genes likely have more than one function. To understand the functional effects of genomic variation, NHGRI launched a consortium called Impact of Genomic Variation on Function (IGVF). genome.gov/news/IGVF
(1/16) Check out our @SimonMJGaudin paper tiny.cc/icuazz from the Canzio lab out in @ScienceMagazine today on how tuning local cohesin trajectories enables differential readout of the clustered Protocadherin (Pcdh) locus across different types of neurons.
Do human RNA-binding proteins have modular regulatory domains that downregulate RNA lifetimes? Our preprint @BintuLab studying this question is now out on bioRxiv, where we indeed identify small domains in RBPs that potently induce RNA degradation! biorxiv.org/content/10.110… (1/9)
Really excited about work led by @jengreitz and @PGuckelberger seeking to harmonize two long-standing & potentially contradictory observations: if cohesin is so important to the formation of chromatin loops, why doesn't degrading it have a bigger impact on gene expression?
I am so excited to share this part of my PhD where we tackle the question: What impact do 3D contacts have on how enhancers regulate their target genes? biorxiv.org/content/10.110… 1/
Exciting news! Jesse (@jengreitz), Marlene (@StanfordBASE), and I have recently received a CIRM grant, and we presumably ranked No. 1 based on the prior year's criteria! I am looking for a talented experimental postdoc to join my team. This position offers close collaboration…
I am so excited to share this part of my PhD where we tackle the question: What impact do 3D contacts have on how enhancers regulate their target genes? biorxiv.org/content/10.110… 1/
Check out ProCapNet by @kellycochra, a robust, local sequence context deep learning model of base-resolution transcription initiation profiles (PRO-cap) that provides deep insights into the cis-regulatory code of initiation & more ... biorxiv.org/content/10.110… 1/
Dissecting the cis-regulatory syntax of transcription initiation with deep learning biorxiv.org/cgi/content/sh… #biorxiv_genomic
Cofactors are a crucial part of the gene activation process. Yet, we still don't have a comprehensive understanding of how and why they are used by different genes. Proud to present our attempt to address this key question, now published nature.com/articles/s4158…
‼️⚠️Registration Deadline TODAY!⚠️‼️ Register now for the 'From Genetic Discoveries to Gene Function in Human Diseases' Conference to secure your slot and join us in Portugal from 13 - 16 July 2024 #GWAS24☀️🏌️ 🔗➡️bit.ly/4bnHuu6
Could not be more delighted to present our work investigating how over 220,000 complex and molecular trait-associated genetic variants affect transcriptional regulation using massively parallel reporter assays! biorxiv.org/content/10.110… See below for a 🧵. 1/n