Bodhi Vani
@bodhivani
ML/statmech/biophys @PrescientDesign. Ex @tiwarylab @IPST, PhD @ Dinner Group @UChicago, @IITB. Likes molecules jiggling and wiggling on manifolds.
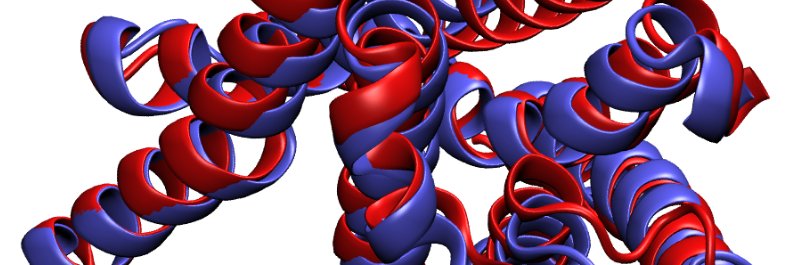
Presenting JAMUN: a model that samples conformational diversity of proteins through Langevin dynamics on a learned smoothed learned manifold and denoising to the physical manifold, demonstrating transferability on a dataset of 2AA. This is just the start! arxiv.org/abs/2410.14621

Come work with me and a bunch of awesome small molecule drug discovery machine learning folks at @PrescientDesign for the summer! roche.wd3.myworkdayjobs.com/ROG-A2O-GENE/j… Looking for PhD students with some experience in ML/physics who can write deployable code.
Only @tiwarylab could come up with this experiment! Go apply and change the paradigm of postdoctoral positions (and tell me how it goes!)
Dive into the world of explainability in graph DL for biomedicine with @_cagarwal and me online at #ISMB2024 on July 8! iscb.org/ismb2024/progr… 🌐💻You'll learn: 🧬 The basics of graph DL & GNNs in biomedicine 💡 Why explainability is key in biomedical ML models 🧵⬇️
OpenFold developed by @gahdritz, @NazimBouatta, and @MoAlQuraishi is a trainable, open-source implementation of AlphaFold2. It is fast and memory efficient, and the code and training data are available under a permissive license. nature.com/articles/s4159…
In (preemptive?) response to all the posts signaling the end of structural biology as we know it
We're delighted that "Protein Discovery with Discrete Walk-Jump Sampling" has won an Outstanding Paper Award at #ICLR2024! 🎉 Congratulations to @nc_frey, @dabkiel1, @karina_zador, Joseph Kleinhenz, Julien Lafrance-Vanasse, Isidro Hötzel, Yan Wu, @stephenrra, @RichBonneauNYC,…
Announcing the #ICLR2024 Outstanding Paper Awards: blog.iclr.cc/2024/05/06/icl… Shoutout to the awards committee: @eunsolc, @katjahofmann, @liu_mingyu, @nanjiang_cs, @guennemann, @optiML, @tkipf, @CevherLIONS
What's a toy model for ligand binding that requires conformational change *for the ligand*?
Internship opportunity at @MSFTResearch #AI4Science on generative protein models. Location Berlin, Amsterdam or Cambridge. Apply here asap: jobs.careers.microsoft.com/global/en/job/…
Ooohhh I highly recommend attending Eric's talk if you can! A rare (ha ha ha) example of learning *science* with ML, not just applications thereof.
Announcing the #ICLR2024 Outstanding Paper Awards: blog.iclr.cc/2024/05/06/icl… Shoutout to the awards committee: @eunsolc, @katjahofmann, @liu_mingyu, @nanjiang_cs, @guennemann, @optiML, @tkipf, @CevherLIONS
We're looking for MD datasets that show protein conformational changes. All-atom or coarse grained, biased or unbiased, sidechains or loops, anything will do. What have we got?
Awesome thread by @tiwarylab about my last first-author work as a post-doc!
Announcing the #ICLR2024 Outstanding Paper Awards: blog.iclr.cc/2024/05/06/icl… Shoutout to the awards committee: @eunsolc, @katjahofmann, @liu_mingyu, @nanjiang_cs, @guennemann, @optiML, @tkipf, @CevherLIONS
Well, we're packed! TIL Nathaniel and my life can be summarized by what I heard one of the movers mutter while taking down notes: "books books books books books boardgames boardgames".

Another AlphaFold residue-index offset hack? 🤯 But this time using it to add disulfide constraints. (neat idea to start with a simple 0,1 matrix, and then using floyd/dynamic-programming to fill in the rest of the offset matrix) biorxiv.org/content/10.110…