Andy Truman PhD
@TrumanLab
Professor in Biological Sciences @unccharlotte Group leader, Charlotte Group for Proteostasis Research Interested in the role of Hsp70 PTMs (Chaperone Code)
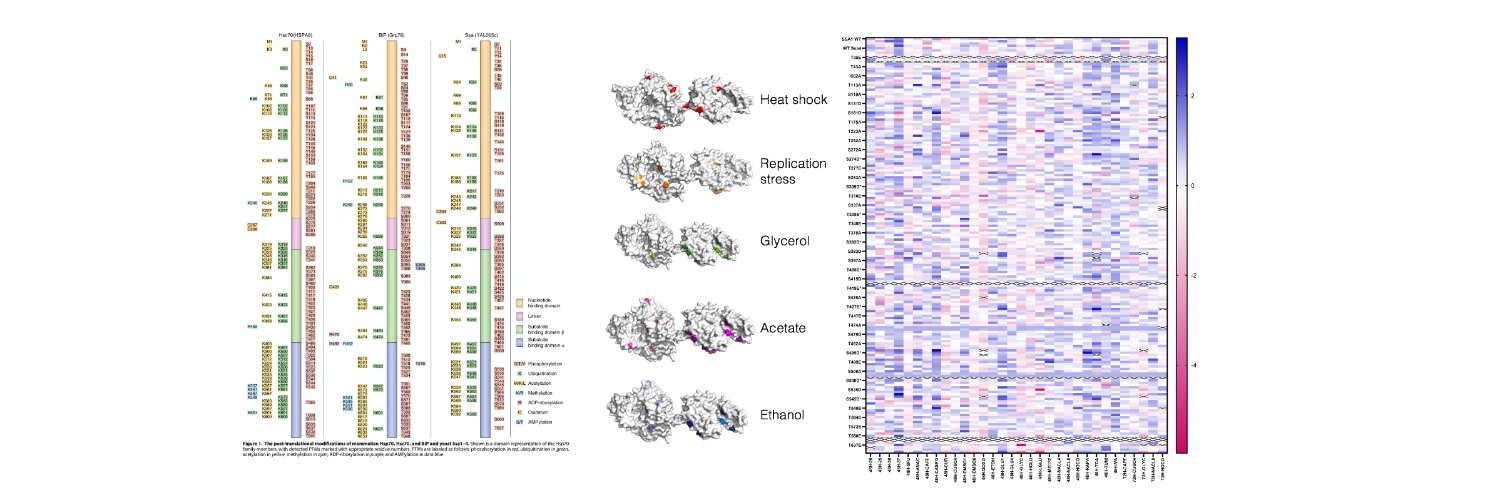
1/Excited to share our latest study "Comprehensive characterization of the Hsp70 interactome reveals novel client proteins and interactions mediated by posttranslational modifications" in @PLOSBiology: journals.plos.org/plosbiology/ar… This work started with a simple idea-
In your opinion, how many first-author publications should a PhD student in biology have to be considered a competitive candidate for a postdoctoral or industry position? @AmerSocVirol @ASMicrobiology @AACR @ASGCTherapy #immunology @UNCCBiology @unccharlotte @UNCCFaculty
Cancer Cell's April's Special Issue is now live! Our central theme: Bridging the gap between foundational cancer research and clinical oncology. Read our Editorial by Editor-in-Chief Steve Mao + an excellent collection of reviews, opinion pieces & articles cell.com/cancer-cell/is…
Two thoughts- 1) if they are not funding at 5% what are they funding and 2) given scoring variation I think we are at the point that the top 25% go into a lottery with winners selected for funding
5 percentile grant, I was suggested to re-submit...which I will, I guess.
NIH will only accept six applications per PI per calendar year. drugmonkey.wordpress.com/2025/07/17/nih… via @drugmonkeyblog
Excited to begin my new role as Associate Chair for Research in Biological Sciences at UNC Charlotte 🔥 @unccharlotte

Great time celebrating the achievements of the lab-graduations, proposal defenses, papers and grants! So grateful for the hard work and energy my team bring 🔥🔥🔥@megan_mitchem @J_Chathura_M @psiddhi273 @DuhitaMirikar @whybee_yb @Rajlekha_061424 @unccharlotte @UNCCBiology

Congratulations to @DuhitaMirikar for presenting her work on multisite Hsp70 phosphorylation fine tuning the DNA replication stress response in yeast! 🔥@UNCCBiology @unccharlotte

Very excited for the 2025 Carolina Symposium on Genome Integrity and Chromaton Regulation @UNCCBiology @unccharlotte @ShanYanLab @ChancellorGaber 🔥🔥🔥

My roman empire-some budding yeast I used to grow: youtu.be/odxvp-3nSw4?si…
⛏️🤙
The biggest reveal this season? Our graduating Norms. 🎓 Congratulations to the students behind the suit who’ve brought the Niner spirit to life! ⛏️
Superb work from @ShorterLab! One of the best papers I have read in a long time 🔥🔥🔥 #chaperones #tdp43 #als #yeast
Scouring the human Hsp70 network uncovers diverse chaperone safeguards buffering TDP-43 toxicity biorxiv.org/content/10.110…
Congratulations to @DuhitaMirikar and @megan_mitchem for passing their proposal defense and to @psiddhi273 on their PhD graduation! So proud of you all!!!! @UNCCBiology @unccharlotte 🔥🔥🔥

Grateful to have been awarded the first UNC Charlotte Klein College of Science award for Research and Scholarship (KNOBEL) 🔥🔥🔥 @UNCCBiology @unccharlotte

Proud to partner with Guilford Technical Community College @_gtcc in offering a seamless transfer option through our #49erNext program to students wishing to pursue a bachelor's degree @unccharlotte. 🤙⛏️💚 ➡️ 49ernext.charlotte.edu
Proud of undergraduate students Shreya Patel and Erica Flores on doing a great job presenting their work at the University of North Carolina at Charlotte Undergraduate Research Symposium! Incredible findings on the Hsp70 and BiP PTM code!


It is finally out! I am incredibly proud to share that my work has officially been published in Cell @CellCellPress! This work was a product of years of dedication and countless hours of research in the labs of Joanna Wysocka and @JaroszLab @StanfordMed cell.com/cell/fulltext/…
Exploring RNA destabilization mechanisms in biomolecular condensates through atomistic simulations | PNAS pnas.org/doi/10.1073/pn…
A great scientific session discussing our new data- #ecDNA #Amplification & #Rearrangements driven by #Epigenetics. & A fun career session @ASBMB Annual Meeting highlighting @CeiFccc & @FoxChaseCancer opportunities! Met many trainees providing the future of discovery and science
Thrilled that our work on this problem with @KarelBrinda, @ZaminIqbal, and others is out in @naturemethods today! We used phylogenetic compression (described in the thread) to compress every microbe ever sequenced onto a flash drive so that it can be searched with a laptop!
So we asked: what sets the fundamental limit on computation on large genomic databases? Evolution! The irreducible entropy in genome collections is bounded by the most parsimonious path to introduce that variability. In other words, optimal compression should echo phylogeny. 4/
Had a fabulous time celebrating our new R1 status and the students and faculty researchers who made it possible! 💚🤙🎊 @unccharlotte