
Genomic Technologies Group
@GenTechGp
Genomic Technologies Group // The Garvan Institute of Medical Research // Australia's leading long-read sequencing service // Enquiries: [email protected]
Nice to play a small part in this. Fantastic work by @htanudisastro & @hdashnow 🧬🧬🧬
Our review article on sequencing & characterising short tandem repeats (STRs) is out in @NatureRevGenet! We delve into short read & long read STR genotyping tools and applications in rare disease & population genetics🧬 go.nature.com/48pWZix @GenTechGp @hdashnow @dgmacarthur
Check out our detailed evaluation of SLOW5 vs POD5 format for raw Nanopore data. TLDR: SLOW5 still the best nanopore data format.
For many of those who were asking on BLOW5 vs POD5 for nanopore signal data, here is a finally detailed benchmark we did: biorxiv.org/content/10.110… Summary: performance of BLOW5 is >= POD5 (from ~= to 100X, see below), with benefit of having ~3 dependencies instead of >50.
Check out the excellent line-up of speakers we have for the @GarvanInstitute Long Read Research Symposium day It is FREE to come along - this November 7th. Reserve your spot eventbrite.com.au/e/garvan-long-…

Not long now until the @GarvanInstitute Long Read Research Symposium on November 7th. We will have talented speakers covering the advantages of both @nanopore and @PacBio data Only a few spots left - don't miss out! 💃 eventbrite.com.au/e/garvan-long-…

Cut your Nanopore raw data files in half with our new compression method ex-zd. Thanks @Hasindu2008 for leading this project 🧬
Introducing ex-zd, a lossless+lossy signal compression for @nanopore signal data. While lossless can only save about 1-3% over vbz, lossy can cut the file sizes by almost half with no noticeable impact on basecalling or methylation calling accuracy. biorxiv.org/cgi/content/sh…
We are hosting a Long Read Research Symposium at @GarvanInstitute ! Our talented speakers will cover the advantages of both @nanopore and @PacBio data. Join us to find out what long reads could do for you 🧬 eventbrite.com.au/e/garvan-long-…

Our #slow5curl paper is out! academic.oup.com/gigascience/ar… Big projects storing hundreds/thousands of @nanopore signal datasets on cloud storage like @awscloud #s3 will be able to save 💸,⏲️ and bandwidth while improving the accessibility of datasets to those with limited computing ..
Excited to share our latest work on somatic mutations in celiac disease! We discover expanded T cell clones with somatic driver mutations in individuals that don't respond to a gluten-free diet. With @FabioItaus, Chris Goodow and many others! Check it out: medrxiv.org/content/10.110…
Super excited to share our work on #celiac disease. We discovered T cells carrying somatic mutations which may explain chronic autoimmune disease medrxiv.org/content/10.110… . Great team work led by @Manu___Singh. Thank you to all authors!
Want an easy to use and fast error/SNP tolerate grep-like tool? Have complex barcodes or indices to demultiplex from raw `omics data? Flexiplex is now published, 🥳academic.oup.com/bioinformatics…, with some great software updates for even more flexibility, github.com/DavidsonGroup/… 1/3
Pleased to report the very first paper from my research group at WEHI is now up on bioRxiv. Flexiplex is a new tool for raw sequencing data that allowed you to identify and error correct barcodes, and can even be used as a general "grep-like" search tool.
Final version of our lamprey epigenome remodeling paper is now out in @NatureComms nature.com/articles/s4146…
Squigualiser preprint with a bunch of usecases is now available 🔥. Your feedback is appreciated. biorxiv.org/content/10.110…
squigualiser has become more powerful 🚀. This signal pileup view of @nanopore r10 DNA signals will deepen your read analysis on IGV. github.com/hiruna72/squig…
Take a break from worrying about the AllOfUs Umap and check out Squigualiser- our new tool for @nanopore signal data exploration. Great engineering by @hiruna72 and @Hasindu2008 as always 🔥🔥biorxiv.org/content/10.110…
Bioinformatics behind: this is an example big project where we used S/BLOW5 ecosystem to efficiently, economically (saving compute, time, energy & money), consistently process & reliably archive 141 nanopore signal datasets (~100TB) on Australia's NCI-Gadi @NCInews supercomputer.
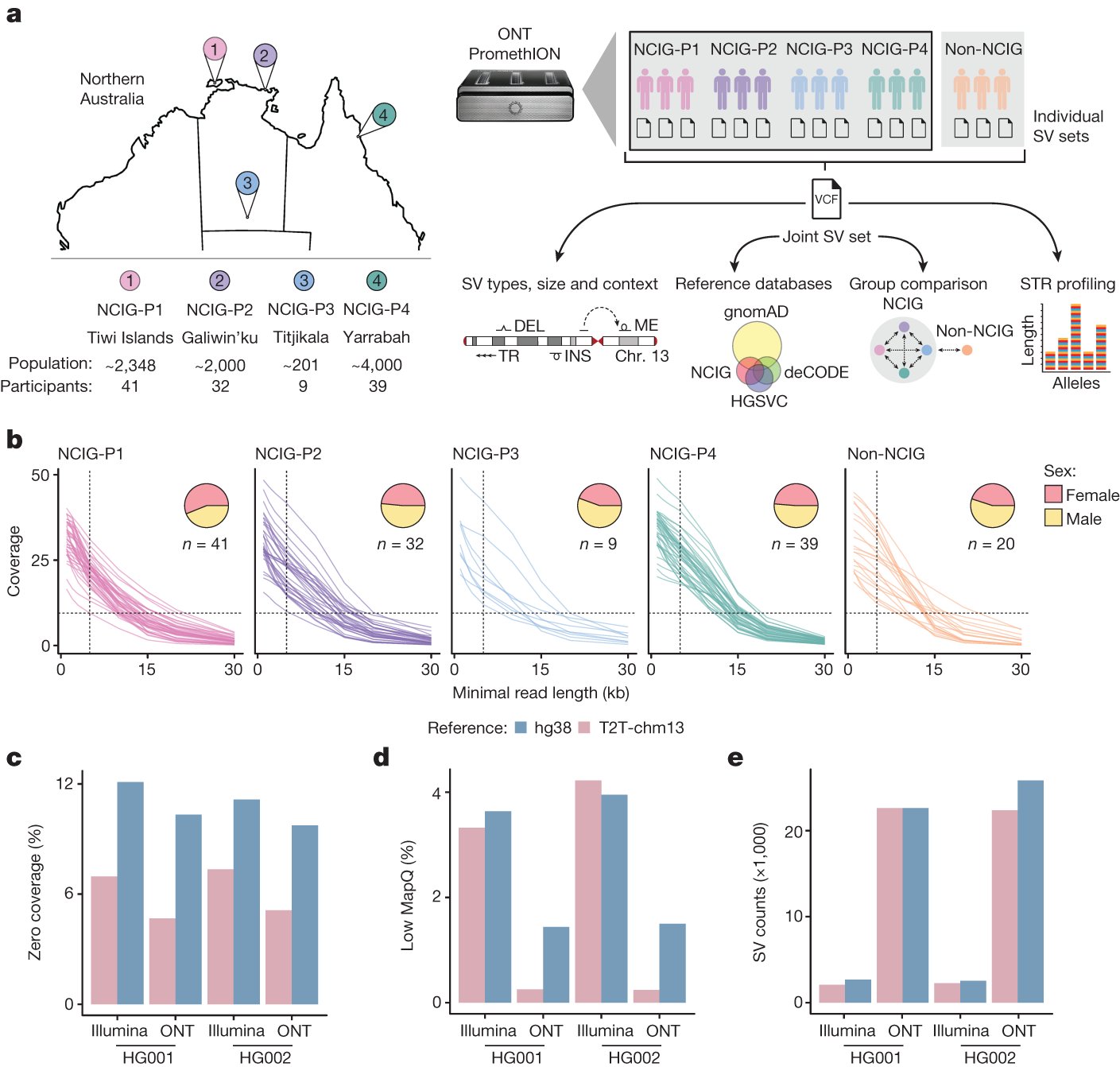
Happy & humbled to see our work on structural variation in Indigenous Australians out in @Nature. Deep gratitude to the Aboriginal communities whose leadership & engagement made it possible, & to our @NCIG2013 collabs @hardiprpatel @azure_peacock 🪃🧬🪃 nature.com/articles/s4158…
Great summary! @BlackOchreOmics @BiancaNogrady “We need to build relationships, we need to prove we are careful & considered stewards … & won’t make moves without communities being across the decisions … & have their voice heard in that process,” nature.com/articles/d4158…
Bunch of legends
Happy & humbled to see our work on structural variation in Indigenous Australians out in @Nature. Deep gratitude to the Aboriginal communities whose leadership & engagement made it possible, & to our @NCIG2013 collabs @hardiprpatel @azure_peacock 🪃🧬🪃 nature.com/articles/s4158…
Happy & humbled to see our work on structural variation in Indigenous Australians out in @Nature. Deep gratitude to the Aboriginal communities whose leadership & engagement made it possible, & to our @NCIG2013 collabs @hardiprpatel @azure_peacock 🪃🧬🪃 nature.com/articles/s4158…
We're fired up about the opportunity to lead this important MRFF-funded project, along with our inspiring collaborators @SusanWhiteAuth1, @mgds26 and many more 🚀🚀🚀
Congratulations to Dr Ira Deveson for being one of the four researchers to receive funding from the Medical Research Future Fund Genomics Health Futures Mission. Learn more: ow.ly/BfSb50QbOfh
Congratulations to Dr Ira Deveson for being one of the four researchers to receive funding from the Medical Research Future Fund Genomics Health Futures Mission. Learn more: ow.ly/BfSb50QbOfh