
Professor Adam Sharples
@DrAdamPSharples
Our Group 1st Demonstrated that Human Muscle possesses an Epigenetic Memory of Exercise -Our DNA remembers exercise! | ExProRugby | Judo
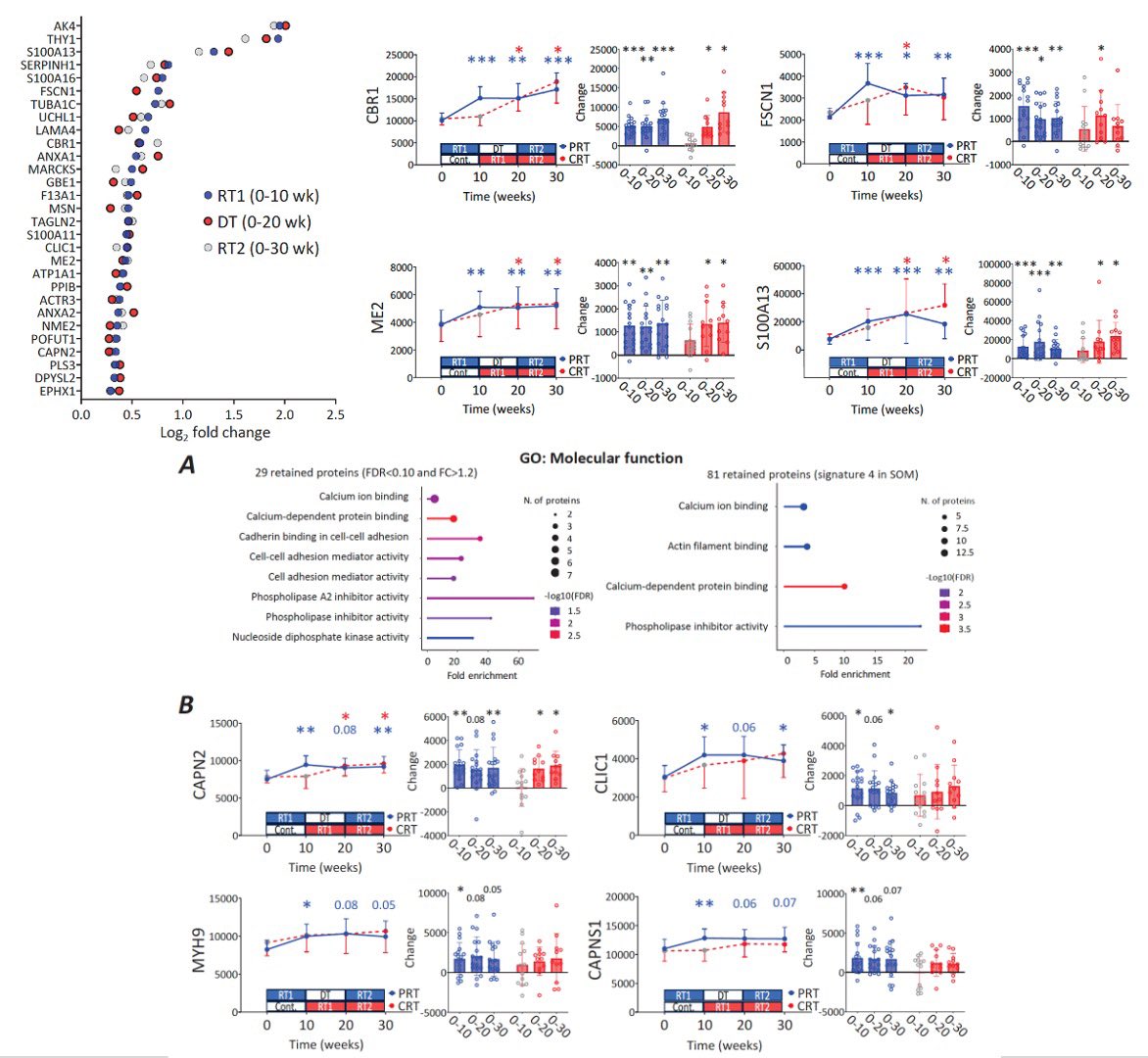
New! @JPhysiol Proteomics of training, detraining & retraining. Reproducible changes in metabolic proteins. Retained changes in cytoskeletal & calcium binding proteins from training remain elevated after detraining Muscle-memory at the protein level! physoc.onlinelibrary.wiley.com/doi/full/10.11…

Fantastic work just just out as a @biorxivpreprint | DNA Methylation Ageing Atlas Across 17 Human Tissues Very honoured to contribute to this work led by @macsuej & @nir_eynon group Very exciting data! biorxiv.org/content/10.110…
Wow! 🤩 if this hold up, it’s is an amazing finding with lots of major implications!
Skeletal muscle stem cell mitochondria are transferred to muscle fibers in response to a hypertrophic stimulus academic.oup.com/function/advan…
Skeletal muscle stem cell mitochondria are transferred to muscle fibers in response to a hypertrophic stimulus academic.oup.com/function/advan…
This is so very much deserved and well overdue! Very excited to see all the UBR and protein quality control mechanisms coming from your lab Dave!
New career chapter 🔓 | Thankful for my mentors and support network after this 10 year post doc journey, which I wouldn’t change for the world 🌎. Excited for all the new challenges ahead 💪🏼. omrf.org/2025/07/17/omr…
Endurance training promotes chromatin accessibility that can be retained! Eluding to other epigenetic modifications that could be involved in muscle memory. It was a pleasure to examine the thesis and pleased to see this finally out in @MolMetab sciencedirect.com/science/articl…
Congratulations Jonathan, Brendan, Nicolas and Julien (and all)! Great to see this fantastic work out in Molecular Metabolism!
Our latest study connects reduced NR4A3 expression with physical inactivity and indicates that NR4A3 downregulation in human muscle has adverse effects on glucose metabolism and protein synthesis. Congrats to @JonoSmith01 @DrBMGabriel @NicoPillon & team! sciencedirect.com/science/articl…
Our latest study connects reduced NR4A3 expression with physical inactivity and indicates that NR4A3 downregulation in human muscle has adverse effects on glucose metabolism and protein synthesis. Congrats to @JonoSmith01 @DrBMGabriel @NicoPillon & team! sciencedirect.com/science/articl…
Amazing to see this collaboration paper in @CellPress @CellReports Molecular landscape of sex- and modality-specific exercise adaptation in human skeletal muscle through large-scale multi-omics integration! Led by @nir_eynon & @MacsueJ cell.com/cell-reports/f…

mTORC1 signalling and protein synthesis are elevated in response to amino acids in human myotubes obtained from young, old, and old trained men link.springer.com/article/10.100…
Thank you @JPhysiol for this honor. Best regards, Juha Hulmi @DrAdamPSharples @Varjosalo_Lab
🎓EDITOR'S CHOICE🎓 Hulmi (@jyu_okl) et al. show that resistance training produces a substantial change in the proteome of skeletal muscle, most of which is reversible. Some proteins remain elevated, implying "muscle memory" of training. 🔗 buff.ly/DkHnDJZ
Congratulations @HornbergerLab and team! This is centre stage and leading the way in deciphering the molecular mechanisms underlying muscle growth with resistance training!
1/10 Excited to write this thread summarizing our new paper in @NatMetabolism, which identifies a potentially critical part of the signaling pathway that drives skeletal muscle growth in response to resistance exercise! Free link rdcu.be/el9am #Myotwitter @NIH_NIAMS
1/10 Excited to write this thread summarizing our new paper in @NatMetabolism, which identifies a potentially critical part of the signaling pathway that drives skeletal muscle growth in response to resistance exercise! Free link rdcu.be/el9am #Myotwitter @NIH_NIAMS
Fantastic news that our article is Editor’s choice in @JPhysiol
Happy that our muscle proteomics article is Editor´s choice in the May issue of 2025 in @JPhysiol physoc.onlinelibrary.wiley.com/doi/10.1113/JP…. @DrAdamPSharples @Varjosalo_Lab
Happy that our muscle proteomics article is Editor´s choice in the May issue of 2025 in @JPhysiol physoc.onlinelibrary.wiley.com/doi/10.1113/JP…. @DrAdamPSharples @Varjosalo_Lab
Absolutely deligthed that David Glass will give the 11th HyperMet talk this Thursday. His title is: "Aging effects of genes effecting skeletal muscle atrophy and hypertrophy" Join on the 15.5. from 15-16 h Munich time tum-conf.zoom-x.de/j/63956266902?… Password: 045026 Please RT
Nice new @biorxivpreprint Efficient Cas9 nuclease-based editing in skeletal muscle via lipid nanoparticle delivery biorxiv.org/content/10.110…
New work from the lab led by the incredibly talented and hard working @JDucharme_PhD and in collaboration with the Esser Lab cell.com/cell-reports/f…
Very excited to see this out in @CellReports after all @MacsueJ and @nir_eynon ‘s excruciating work!
Congratulations to our talented postdoc @MacsueJ , who had her ‘multiome exercise’ paper accepted @CellReports Macsue analysed more then 1000 muscle samples to identify multi- molecular signals of exercise both aerobic and strength Preprint: biorxiv.org/content/10.110…