Daniel Figeys
@DanielFigeys
University of Ottawa. Microbiome, metaproteomics, metabolomics, metagenomics, bioinformatics, drug- microbiome interactions, precision microbiome nutrition
Huge thanks to all the collaborators at University of Lille (Dr. Trottein team), University of Ottawa (Dr. Figeys team @DanielFigeys), Health Canada (Dr. Sean Li team), and University of Campinas (Dr. Vinolo team).
An article published in @MicrobiomeJ presents PhyloFunc: a novel functional beta-diversity metric that incorporates microbiome phylogeny to inform on metaproteomic functional distance. microbiomejournal.biomedcentral.com/articles/10.11…
Vacancy! We are looking for a Group Leader in Functional #Microbiome Research to join the Quadram Institute. Amazing start-up and support. Fantastic translational facilities. This is the place to be, check it out. quadram.ac.uk/vacancies/grou…

Today marks my last day at @uOttawa. Grateful for the years of collaboration, and growth. From founding the OISB to launching the School of Pharmaceutical Sciences—what a journey. Thank you to all who were part of it and to my amazing lab. On to the next chapter! 🚀 @NorthOmics
Metaproteomic network. Sun et al. analyzed peptide correlations from a large-scale #metaproteomics study. Results: better coverage at genome levels and new links between known and uncharacterized proteins. @NorthOmics biorxiv.org/content/10.110…

🚨 We mapped 4.6M #microbial protein responses to 312 compounds, uncovering 3 distinct functional gut #microbiome states and revealing how neuropharma triggers deep shifts (lower functional redundancy, higher AMR proteins). @li_leyuan @NorthOmics biorxiv.org/content/10.110…
PhyloFunc: A new beta diversity metric considering evolutionary relationships and protein expression. Check out the video. @li_leyuan @NorthOmics researchsquare.com/article/rs-601…
Interested in studying the functions of the microbiome?Check out PhyloFunc, a novel functional beta-diversity metric that incorporates #microbiome phylogeny to inform on #metaproteomic functional distance. @NorthOmics microbiomejournal.biomedcentral.com/articles/10.11…
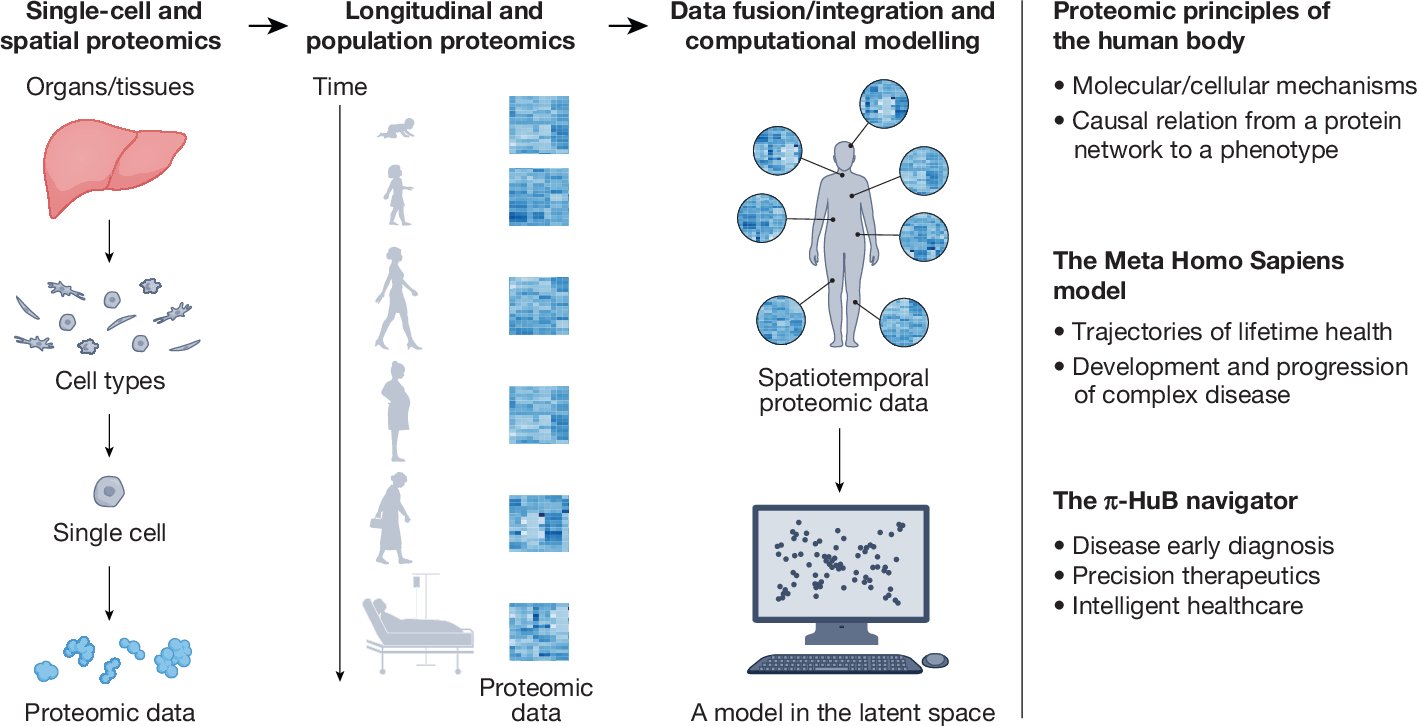
Excited to be part of the proteomic navigator of the human body (π-HuB). This will transform our understanding of biological processes in human, including the human gut #microbiome . nature.com/articles/s4158… @NorthOmics
A machine learning system that simulates proteins with more than 10,000 atoms with quantum accuracy. nature.com/articles/s4158…
Delighted to share that I’ll be joining the @QuadramInstitute as Director & CEO in April 2025! I am honoured to work with an incredible team and partners to push the boundaries of gut health, microbiology, and food science research.
🆕 We are delighted to announce the appointment of Professor @DanielFigeys as our new Director and Chief Executive. buff.ly/40H5BRN
Interested to do #metaproteomics using DIA mass spectrometry, while being free from generating DDA results? Check out the new version of MetaLab-DIA imetalab.ca. #FreefromDDA @hyrnzb @NorthOmics
MetaLab Platform Enables Comprehensive DDA and DIA Metaproteomics Analysis biorxiv.org/content/10.110… --- #proteomics #prot-preprint
MetaLab Platform Enables Comprehensive DDA and DIA Metaproteomics Analysis biorxiv.org/cgi/content/sh… #biorxiv_bioinfo
Gut bacteria identified which can degrade IgA. Read more here science.org/doi/10.1126/sc… and in this Perspective science.org/doi/10.1126/sc…
Out now in @ScienceAdvances: Antibiotics damage the colonic mucus barrier in a microbiota-independent manner @Mucus_Man @meytaln science.org/doi/10.1126/sc…
In situ targeted base editing of bacteria in the mouse gut | Nature nature.com/articles/s4158…