
Blainey Lab
@BlaineyLab
Official Twitter account of the Blainey Lab @broadinstitute
Recently published in @PNASNews our high-throughput screen of 1.3M+ antibiotic combos identified P2-56 as a rifampin potentiator against A. baumannii & K. pneumoniae. pnas.org/doi/10.1073/pn…
New @biorxivpreprint! w/ our high-throughput screening approach, screened 1.3M+ antibiotic combos against high priority, drug-resistant bacterial pathogens, identified small molecule P2-56 as a potentiator of rifampin against A. baumannii and K. pneumoniae biorxiv.org/content/10.110…
Using an image-based screening method, Broad and Boston University researchers found potential new drug targets for reducing Ebola infection severity, uncovering human genes that, when silenced, impaired Ebola’s ability to replicate. broad.io/Ebola-News-0724
Out now in @NatureMicrobiol @beccajcarlson and our lab collab w/ Rob Davey's lab @neidl on high-content image-based perturbational screening for regulators of Ebola virus infection @broadinstitute nature.com/articles/s4156…

We’re @BlaineyLab sharing a major update to CROPseq-multi, our versatile system for CRISPR screens that is compatible with individual and combinatorial perturbations, diverse SpCas9-based technologies, and multiple high-content, single-cell readouts. biorxiv.org/content/10.110…
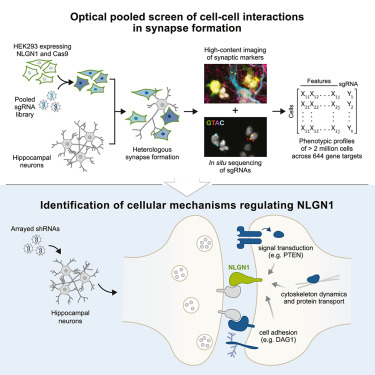
Out now in @CellReports our collab w/ @BiedererLab on image-based pooled genetic screening to identify regulators of synapse formation cell.com/cell-reports/f…
Researchers at the @broadinstitute have created a pipeline for discovering unique combinations of molecules that increase the effectiveness of #antibiotics against drug-resistant bacteria. ✏️ Abstract: bit.ly/4kry8lu 📰 Press Release: bit.ly/3T5TSap
Excited to share my primary PhD work! Vibrio isolates can produce peptide deformylase inhibitors that induce prophages in their lysogenic competitors. Not only is it SOS-independent, but also serves as the first example linking translational stress with prophage induction 👀 1/
Bacterial peptide deformylase inhibitors induce prophages in competitors biorxiv.org/content/10.110… #biorxiv_micrbio
Excited to share the 1st of several stories dissecting TEs + chromatin in normal/malignant hematopoiesis. We identified histone demethylases KDM4A/C as vulnerabilities in DLBCL + discovered novel KDM4 inhibitors. Preprint👇 @broadinstitute @BostonChildrens biorxiv.org/content/10.110…
Excited to see the work from @mgentili_ @beccajcarlson and others from our collab with the Hacohen lab @broadinstitute out now!
Our image-based genome-wide screen identifying new regulators of STING localization is now online at @CellSystemsCP! authors.elsevier.com/c/1kEhc8YyDfql…
In collaboration with Reuben Saunders, @JswLab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! biorxiv.org/content/10.110…
In collaboration with @weallen1 and Xiaowei Zhuang, we have developed Perturb-multi: biorxiv.org/content/10.110…
In collaboration with Reuben Saunders, @JswLab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! biorxiv.org/content/10.110…
Available now via @biorxivpreprint is our preprint on live-cell transcriptomics with engineered virus-like particles @broadinstitute biorxiv.org/content/10.110…
Out now in @cellcellpress - our story about metabolic differences in the vaginal microbiome led by @mzhu_ology @broadinstitute in collaboration with the @kwonlab at @ragoninstitute and others: cell.com/cell/fulltext/…

We're hiring for a postdoc to work with us on the technical development and deployment of optical pooled screening (OPS) technology! Details and apply here: broadinstitute.avature.net/en_US/careers/…
We're hiring! We are seeking an exceptional postdoc associate to help us work on solving the antibiotic resistance crisis, details and apply here: broadinstitute.avature.net/en_US/careers/…
Now out on @biorxivpreprint! Thank you to our alum @beccajcarlson for this great Tweetorial! @broadinstitute
I’m excited to share a new manuscript from my PhD! doi.org/10.1101/2024.0… We performed a genome-wide optical pooled screen (OPS) and identified hundreds of novel Ebola virus regulators.
We are excited to share our new @biorxivpreprint about CROPseq-multi with the community, many thanks to our @russelltwalton for this Tweetorial about his work with @Yue_Qin_SysBio! @broadinstitute @MITdeptofBE @Schmidt_Center
Our new preprint describes “CROPseq-multi”: a versatile solution for multiplexed perturbation and decoding in pooled CRISPR screens doi.org/10.1101/2024.0… Reagents on Addgene: addgene.org/browse/article…
Excited to share our preprint on metabolic differences in the vaginal #microbiome. We find key differences in fatty acid metabolism among #Lactobacillus species & leverage those findings to identify a potential new therapy for bacterial vaginosis (BV) & #WomensHealth 🧵 (1/n)
Vaginal Lactobacillus fatty acid response mechanisms reveal a novel strategy for bacterial vaginosis treatment biorxiv.org/cgi/content/sh… #bioRxiv
And check out the fantastic MIT News piece by Anne Trafton about our STING paper news.mit.edu/2023/study-fin…
