William Allen
@weallen1
Biology of homeostasis & resilience. New tools to map + control cells. Asst prof @StanfordDevBio. @Harvard Junior Fellow, neuro Ph.D. @Stanford, MPhil @MRC_LMB
Very excited to announce that I'll be starting a laboratory studying the biology of homeostasis as an assistant professor in the Department of Developmental Biology (@DevBioStanford) at @Stanford in July! allenlabstanford.org

My paper is out!! 🎉 "Opponent control of reinforcement by striatal dopamine and serotonin", @Nature Here, we show that dopamine and serotonin signals form a gas-brake system for reward in the mammalian brain 🧠 THREAD ⬇️ 1/n
Excited that the Zhuang Lab is now on X!
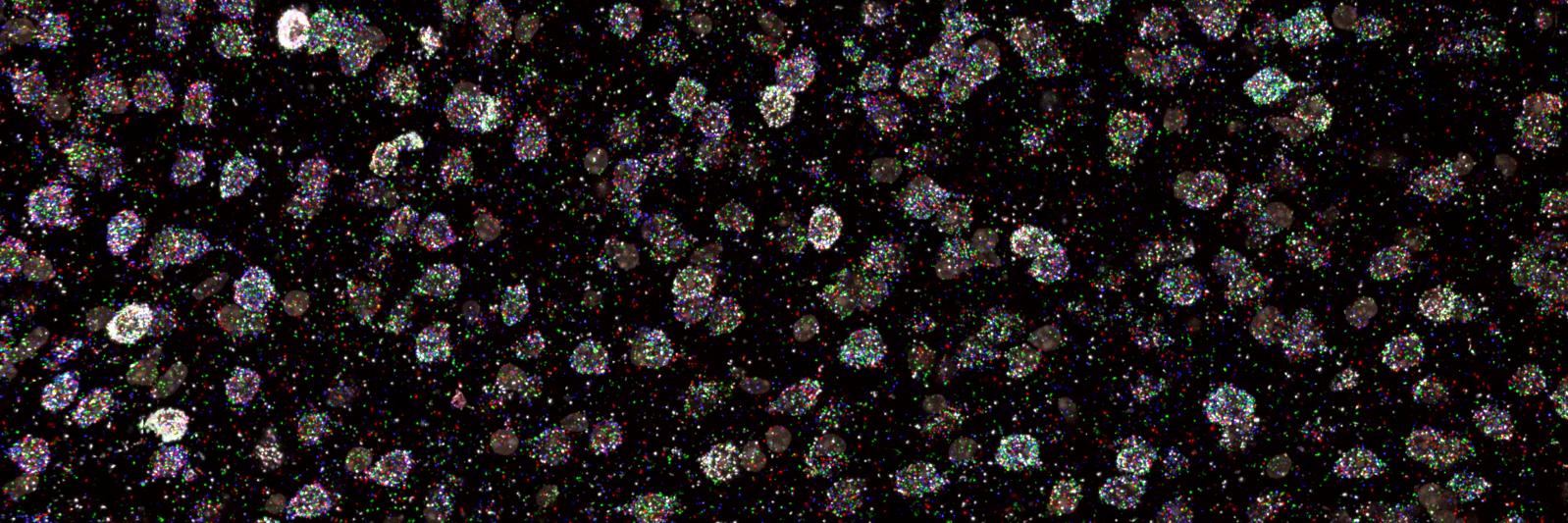
Check out our preprint on Perturb-multi (doi.org/10.1101/2024.1…), a multi-year collaborative effort that enabled multi-modal in vivo genotype-phenotype mapping by pooled genetic perturbations and high-dimensionality phenotype readout by imaging and sequencing.
Check out our preprint on Perturb-multi (doi.org/10.1101/2024.1…), a multi-year collaborative effort that enabled multi-modal in vivo genotype-phenotype mapping by pooled genetic perturbations and high-dimensionality phenotype readout by imaging and sequencing.
In collaboration with Reuben Saunders, @JswLab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! biorxiv.org/content/10.110…
Delighted to share this preprint from Prof. Rong Fan Lab @RongFan8 and Prof. Sidi Chen lab @sidichen on Spatially Resolved in vivo CRISPR Screen Sequencing via Perturb-DBiT. Congatulations, Alev, Xiaolong, Feifei, Paul, Sidi and Rong!
Thrilled to share Perturb-DBiT — spatial unbiased in vivo perturb-seq with genome-scale CRISPR libraries! Hope you enjoy reading it over the Thanksgiving week 🥰 Kudos to @AlevBaysoy, Xiaolong, Feifei, and Paul. & collaboration with @sidichen, Hongbo Chi. biorxiv.org/content/10.110…
Thrilled to share Perturb-DBiT — spatial unbiased in vivo perturb-seq with genome-scale CRISPR libraries! Hope you enjoy reading it over the Thanksgiving week 🥰 Kudos to @AlevBaysoy, Xiaolong, Feifei, and Paul. & collaboration with @sidichen, Hongbo Chi. biorxiv.org/content/10.110…
delighted to share this exciting news with Prof. Xiaowei Zhuang @zhuanglab and Prof. Jonathan Weissman @JswLab, on their latest preprint reporting Perturb-Multi: a multimodal in vivo pooled CRISPR screen 🤩 congrats Reuben and William @weallen1 👏🍾 biorxiv.org/content/10.110…
New work from @arcinstitute on the cover of Science this week!
A new Science study presents “Evo”—a machine learning model capable of decoding and designing DNA, RNA, and protein sequences, from molecular to genome scale, with unparalleled accuracy. Evo’s ability to predict, generate, and engineer entire genomic sequences could change the…
Check out the preprint describing Perturb-Multi from Arc Ignite Awardee @weallen1, Reuben Saunders (now at Arc), @JswLab and Xiaowei Zhuang. A general and scalable approach applied here to mouse liver, Pertub-Multi generates rich, multimodal data that can generate fundamentally…
In collaboration with Reuben Saunders, @JswLab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! biorxiv.org/content/10.110…
Happy to share the lab’s latest research tagging psychedelic-activated neurons in mice. See the post below from first author @jessie_muir for a tweeprint on the work. Thanks to wonderful collaborators @DEOlsonLab @LinTianPhD and to our reviewers/editor! science.org/doi/10.1126/sc…
Excited to share the Kim Lab’s newest paper in @ScienceMagazine with Sophia Lin (twitterless) and @tinakim_neuro ! We look at circuit mechanisms of psychedelic action underlying therapeutic-like effects in the PFC. science.org/doi/10.1126/sc…
One key component of Perturb-Multi is in vivo Perturb-seq of formaldehyde-fixed tissue, enabled by the Flex reagents from @10xGenomics.
In collaboration with Reuben Saunders, @JswLab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! biorxiv.org/content/10.110…