Valentina Boeva
@val_boeva
Assistant professor at the Department of Computer Science
#AACRai25 cochairs Valentina Boeva and Benjamin Haibe-Kains adjourned the AACR Conference on Artificial Intelligence and Machine Learning. We thank all of the chairs—including @MarzyehGhassemi and @BoWang87—for developing an outstanding program. @val_boeva @bhaibeka
🔥Just wrapped up an incredible three days in Montreal at the AACR (@AACR ) Special Conference on Artificial Intelligence and Machine Learning — the first AACR event fully dedicated to exploring how AI is reshaping cancer research and care. I was deeply honored to serve as one of…
Congratulations to the team on the milestone paper!
*New Open-Access Long Read Resource*. We sequenced 1,019 genomes from the 1000 Genomes Project sample cohort using @nanopore. Sequencing data is available at bit.ly/4m8dlE2. @embl @HHU_de @IMPvienna @CRGenomica nature.com/articles/s4158… [1/8]
Thank you HiTSeq for inviting me to talk to the wonderful #ISMBECCB2025 audience about our work! UniversalEPI for Hi-C predictions biorxiv.org/content/10.110… ASAP for ATAC-seq predictions biorxiv.org/content/10.110… Also, come to visit Aayush Grover’s poster on Thursday!…
We have officially started #HitSeq track @hitseq #ISMBECCB2025. Francisco de la Vega introduces our first #keynote speaker, Valentina Boeva, with her talk "Learning variant effects on chromatin accessibility and 3D structure without matched Hi-C data"
🚨 We're hiring! @NCMBMnews is looking for 2 new group leaders to join our vibrant research community. Ready to build your own group with strong support and a solid startup package? Interested and attending #ISMBECCB2025 @iscb? Let’s connect! 🔗 jobbnorge.no/en/available-j…
Good bye Vancouver, hello Liverpool #ISMBECCB2025 ! Hope to see you there!

An #ICML2025 dinner with the smartest and most handsome conference delegates :-)

Do current genomic language models (pre-trained on whole genomes) learn a foundational understanding of biology in the non-coding region of human genomes? A new evaluation led by @AmberZqt suggests not yet! 1/N paper: biorxiv.org/content/10.110…
Join our lab's presentations at ICML'2025 @icmlconf in beautiful Vancouver! 1. Thursday, Olga Ovcharenko (@o_ovcharenko) will present our work with @sscdotopen and @vogt_je on "scSSL-Bench: Benchmarking Self-Supervised Learning for Single-Cell Data", selected for a spotlight…

On Thursday, Olga will present her research on "scSSL-Bench: Benchmarking Self-Supervised Learning for Single-Cell Data". This paper is joint work with ETH Zuerich and was selected as a spotlight poster: icml.cc/virtual/2025/p… (2/3)
Goodbye Montreal! The special #AACR conference on AI and ML in Cancer Research was a fantastic event! Hope it’s not the last time we organize it! #AACRai25 @bhaibeka


Alexander Tropsha talking about challenges of drug discovery in cancer research at #AACRai25 - so much to do to fill the gap!

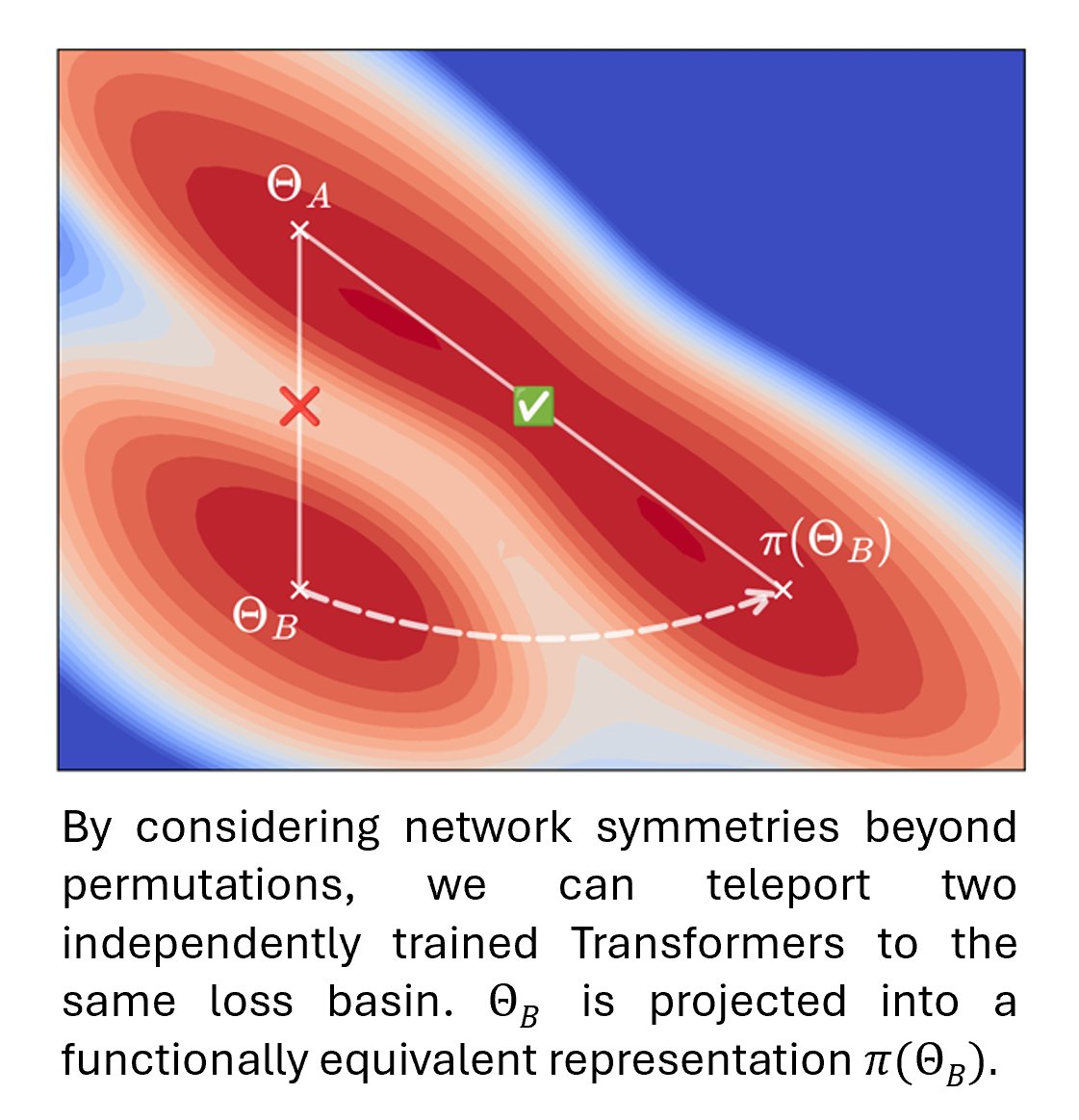
1/ 🚨 New paper alert! 🚨 We explore a key question in deep learning: Can independently trained Transformers be linearly connected in weight space — without a loss barrier? Yes — if you uncover their rich symmetries. 📄 arXiv: arxiv.org/abs/2506.22712
🔥A very cool piece from Alexander Theus and team is finally on arXiv: arxiv.org/abs/2506.22712 Ever wondered if you can linearly interpolate between transformers and still get a working model? Now you'll know how. A breakthrough! 🚀
Congratulations on the successful PhD defense, Dr. Nikita Janakarajan @niklexical ! Amazing presentation! 🚀Best of luck with your future career! 🎊🥳


Our lab at the Weizmann Institute was hit tonight by Iran We will rebuild and return 💪