
HiTSeq 2024
@HiTSeq
HiTSeq is to be held in Montreal, Canada on July 13-14, 2024 where we will discuss the latest advances in computational techniques for analysis of HTS data.
Meet our second sponsor, Oxford Nanopore @oxfordnanopore at @HiTSeq track #ISMBECCB2025. During his talk, Mike Vella takes us to the latest applications of machine learning methods to tackle one of the most difficult challenges for long-read sequencing: base calling

We open our second day at @HiTSeq with our last keynote speaker, Tobias Marschall, and his insightful talk on the human pangenome and the challenges of structural variation analysis. His work has focused on tackling current limitations, such as sample size and remaining gaps

Meet our amazing sponsor, PacBio @PacBio for @HiTSeq track at #ISMBECCB2025 represented by Elizabeth Tseng with her talk "Bioinformatics analysis for long-read RNA sequencing: challenges and promises" #iscb #sequencing #applications #Liverool #UK
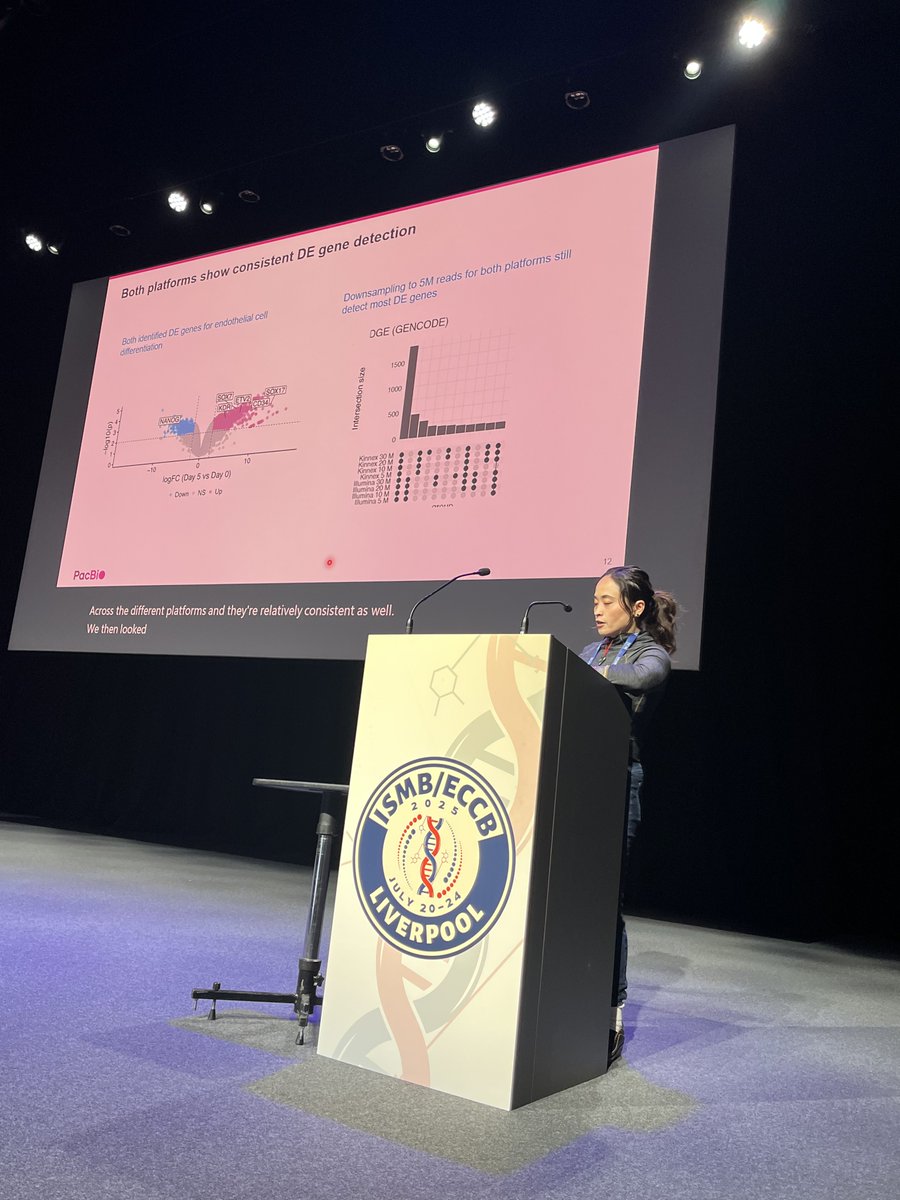
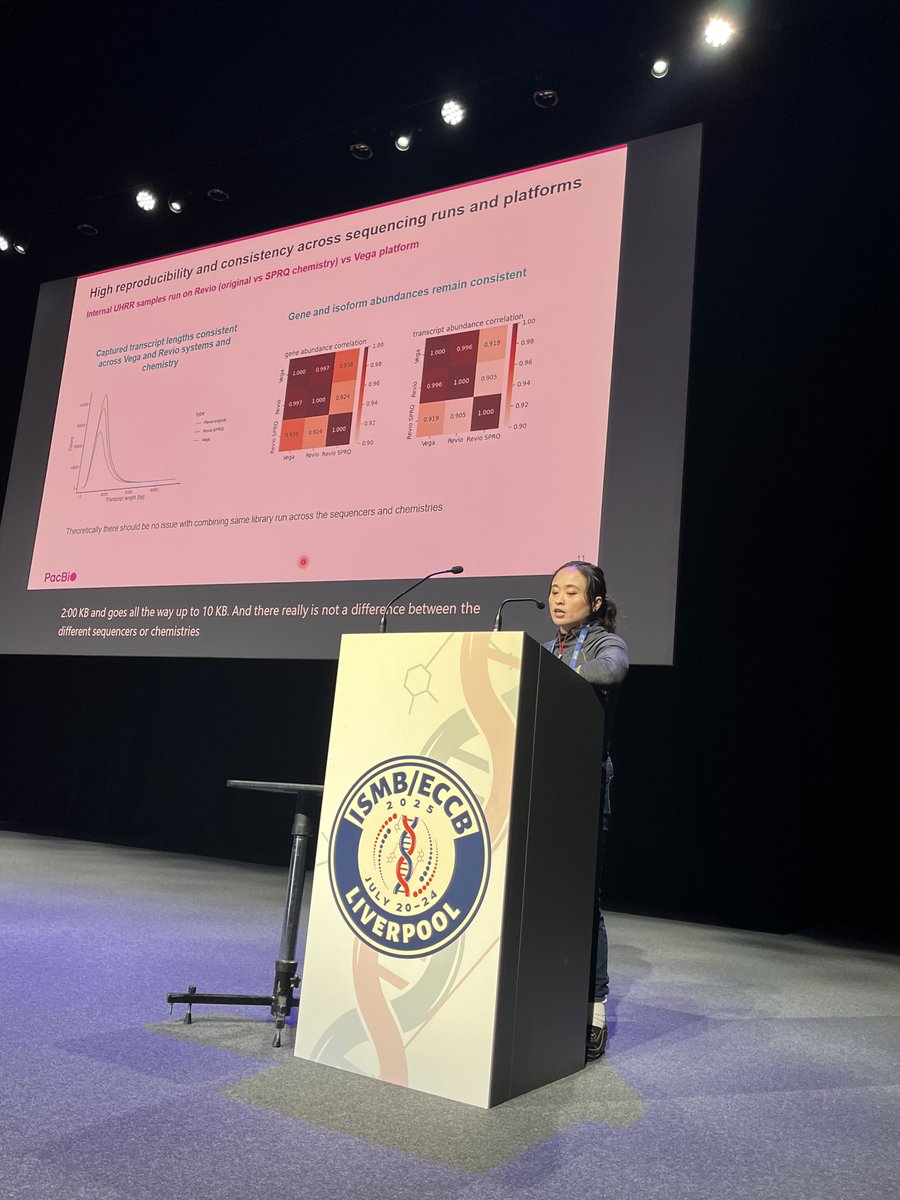
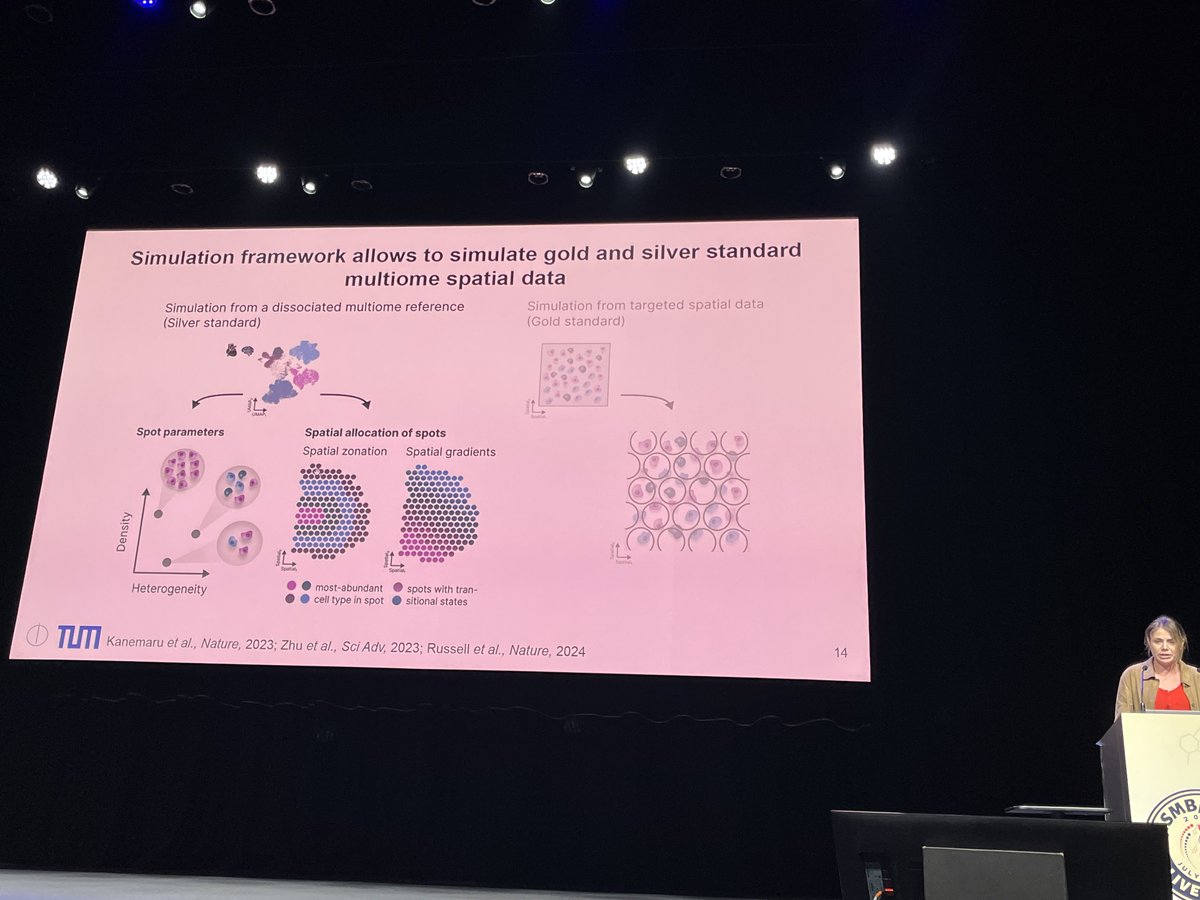
Fantastic first #proceeding talk at @HiTSeq by Lauren Martens! In her work, she benchmarks different tools to perform deconvolution on spatial data using single-cell data references to estimate cell-type fractions in a tissue context! #transcriptomes#algorithms

We have officially started #HitSeq track @hitseq #ISMBECCB2025. Francisco de la Vega introduces our first #keynote speaker, Valentina Boeva, with her talk "Learning variant effects on chromatin accessibility and 3D structure without matched Hi-C data"


We are thrilled to have our @HiTSeq member, Dr. Christina Boucher, featured in a new episode for the #BioinfoBridges @iscbsc #podcast! 🎙️🔬 Have you ever wondered how bioinformatics is used to decode microbial genomes? 🚀 Full Episode Now! 🎙️ Spotify: 🔗open.spotify.com/episode/7uhmP3…
#ISMB2024 Time to leave the football and come back to the last session of #HiTSeq2024 in room 517d - four exciting talks streaming now!
#ISMB2024 come to room 517 to hear about the applications of quantum computing in genomics! #HitSeq2024
#HitSeq2024 is starting again in room 517d - more talks on basic algorithms for sequence analysis #ISMB2024
We open our second @hitseq session track at #ISMB2024 with our fantastic third keynote speaker, Kin Fai Au, discussing methods of using long reads to improve gene isoform quantification #algorithms #genomics


HitSeq2024 is starting now at room 517d with a great keynote by Kin Fai Au: “Why and how long reads are used to improve gene isoform quantification” - join us! #ISMB2024
TODAY at 5 pm, @HiTSeq will have its fantastic second keynote speaker, Evan Eichler, at #ISMB2024 Room 517d! He aims to understand the evolution, pathology, and mechanism of recent gene duplication and DNA transposition within the human genome. Looking forward to seeing you!

We are proud to have @PacBio sponsor the @HiTSeq track at #ISMB2024. As a global leader in advanced #sequencing technologies, their mission is to enable the promise of #genomics to better human health.

#ISMB2024 We open the @HiTSeq track with our wonderful first keynote speaker, Sushmita Roy @sroyyors, enlightening us about how changes in 3D genome organization lead to transcriptional changes and unsupervised learning methods for regulatory genomics! #algorithms #genomics


🚨Do you analyze sequencing data? Then @HitSeq COSI is for you! You are still in time to submit your work to the #ISMB2024 conference here: iscb.org/ismb2024/submi… Check our webpage for more info: hitseq.org #sequencing #analysis #genome #reads #methods #tools

Looking at the #ISMB2024 submission site of @iscb and not knowing which COSI to choose? If your work involves the analysis of sequencing data, just choose @HitSeq. Then you are good… iscb.org/ismb2024/submi…
Ready to present your computational work to the world? You are still on time to submit your @HitSeq #ISCB COSI abstract to the biggest bioinformatics conference of the year #ISMB2024, here 👉 iscb.org/ismb2024/submi… DEADLINE APRIL 19TH #talk #ISCB #sequencing #algorithms