
Steve McCarroll
@s_mccarroll
Variation makes biology rich and interesting – let's figure out how it works
Excited to share this discovery: Huntington's disease is a DNA process for almost all of a cell's life. Inherited HD alleles are innocuous, just unstable – CAG repeats slowy expand throughout life, acquiring toxicity only when quite long (>150 CAGs). biorxiv.org/content/10.110…
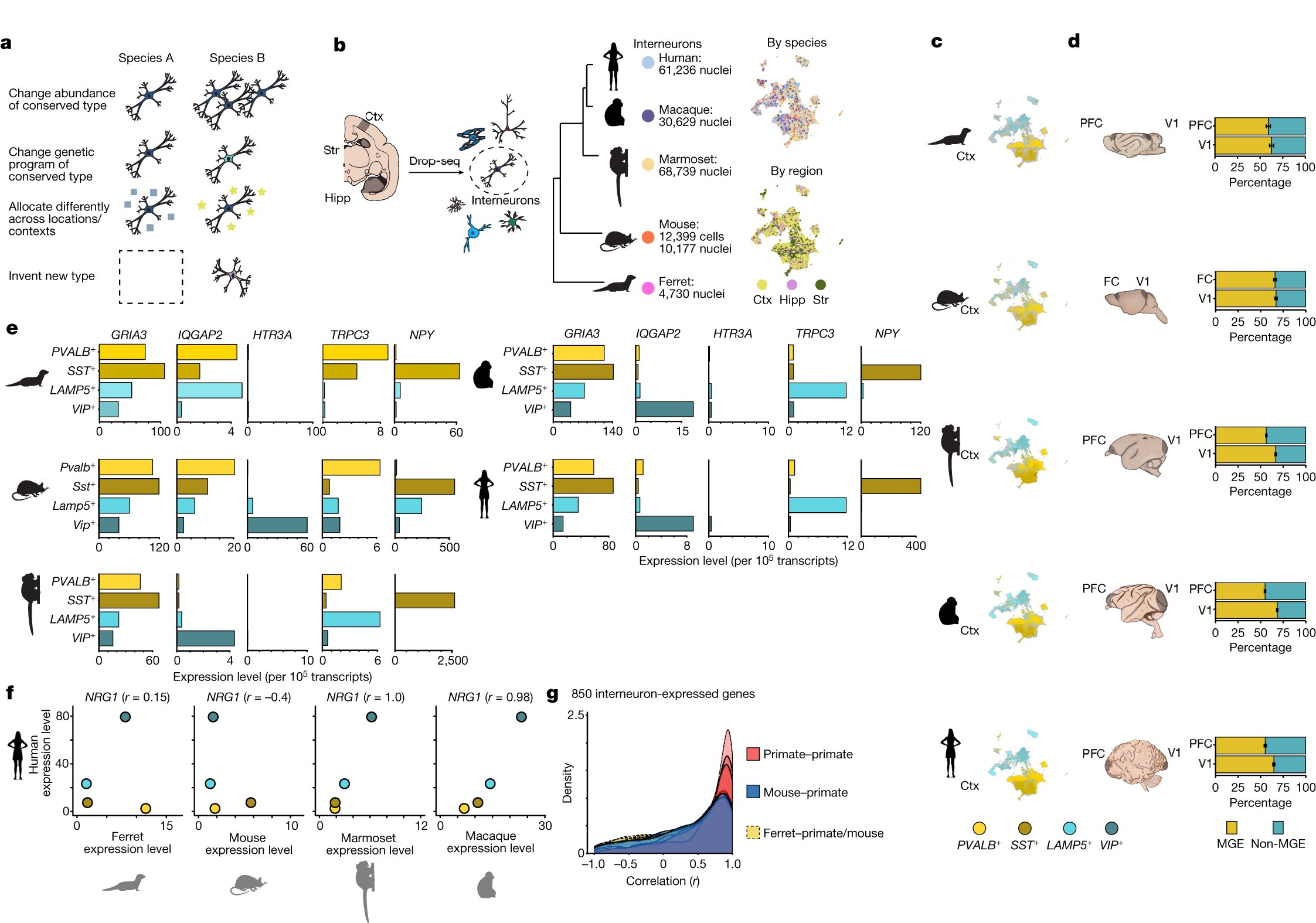
Remarkable evolutionary innovations by primates. My favorite: a third of the interneurons in the striatum are a novel type without homolog in mouse or ferret. Excited to share this work from team led by @fennamk with @GordFishell @guoping_feng nature.com/articles/s4158…
Excited to share the paper: Huntington's disease is a DNA process for almost all of a cell's life. Inherited HD alleles are innocuous, just unstable – CAG repeats slowy expand throughout life. We call it a "ticking DNA clock". cell.com/cell/fulltext/…
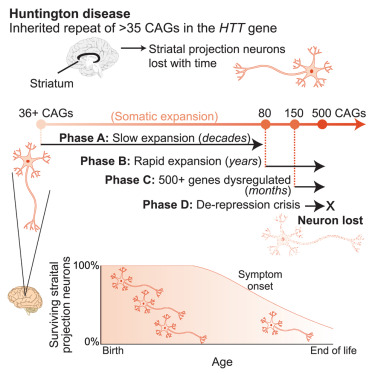
This is the type of amazing #ASHG23 plenary talk like when triplet repeat disorders or BRCA1 were discovered. Robert Handsaker of MGH finding that only specific neurons expand CAG repeat further in patients with Huntington as they age!!
Marmosets have their siblings' microglia – not kidding about this: biorxiv.org/content/10.110…
To learn how noncoding, polygenic variation shapes phenotypes, we need to find ways to think outside of the enhancer box. Excited to share this effort to do that, work with @danweiner92 @eliserobinson. Check out Dan’s tweetorial! ()
Excited to share our latest work!! We uncover functional convergence of common+rare genetic risk factors for autism around 16p11.2, a region of longstanding mystery in neuropsych research 🧠 🧵 👇 (1/14) medrxiv.org/content/10.110…
Excited to share our latest work!! We uncover functional convergence of common+rare genetic risk factors for autism around 16p11.2, a region of longstanding mystery in neuropsych research 🧠 🧵 👇 (1/14) medrxiv.org/content/10.110…
Viral pandemics in "cell villages": reveals strong effect of a common protective human genetic variant. Collaboration with @EgganLab @mfwells5 , toward new ways of learning how human genetic variation shapes cell biology. biorxiv.org/content/10.110…
Just out online in Science: our paper together with Po-Ru Loh's lab on the surprisingly large effects that common VNTRs have on many human phenotypes. Common variants with large effects, hiding in plain sight! science.org/doi/10.1126/sc…
Excited to share this technology for studying cells' synaptic connections alongside their RNA expression. Work led by @your_arpy, now a professor at the Vollum Institute: biorxiv.org/content/10.110…
Common variants with large effects on phenotypes – hiding in plain sight! Here we developed ways to analyze VNTRs in large-n human genetics. The strengths of the associations – and what they revealed about genetics and protein domains – astonished us. biorxiv.org/content/10.110…