
Romero lab
@romerolab1
Romero Lab @DukeUBME. Run by his grad students. We love proteins and engineering them.
🎉Congrats to Nate for his preprint, jointly with @vatsanraman, where training on multifunctional data enabled precise control over designing phages with complex infectivity & specificity. Big shifts in host targeting come from just a few mutations! tinyurl.com/yc4wtn8h
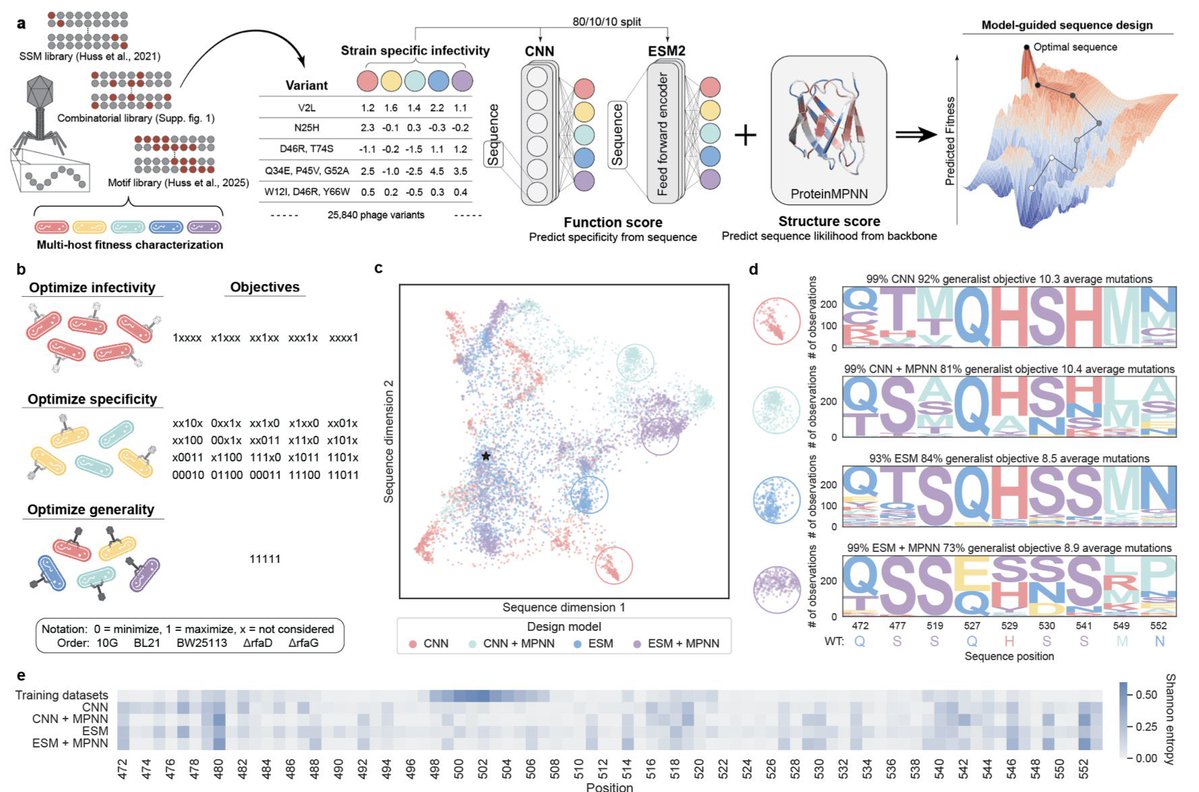
Designing proteins for multiple, often competing objectives is a major challenge. In our new preprint, we use deep learning + protein language models to design phages with higher activity, defined specificity, and generalizable infectivity—meeting 26 design goals across several…
Multiobjective learning and design of bacteriophage specificity 1.This study presents a landmark demonstration of multiobjective machine learning to design bacteriophages with tunable infectivity, precise host specificity, and broad generality, optimizing over 26 complex tasks…
Excited to share @NathanielBlalo2 and @srivsesh preprint teaching protein language models to generate beyond what evolution has explored. They introduce Reinforcement Learning with eXperimental Feedback (RLXF) to steer generation toward enhanced and non-natural functions.
Reinforcement learning with experimental feedback (RLXF) shifts protein language models so that they generate sequences with improved properties. @NathanielBlalo2 @srivsesh @romerolab1
Functional alignment of protein language models via reinforcement learning 1.This work introduces RLXF, a general framework for aligning protein language models (pLMs) with experimental functional objectives using reinforcement learning—enabling the generative design of…
Functional alignment of protein language models via reinforcement learning 1.This work introduces RLXF, a general framework for aligning protein language models (pLMs) with experimental functional objectives using reinforcement learning—enabling the generative design of…
We are excited at the @romerolab1 to share our new preprint introducing RLXF for the functional alignment of protein language models (pLMs) with experimentally derived notions of biomolecular function!
RLXF is on @biorxivpreprint! We introduce a PPO-based workflow to align the logits of any protein language model away from evolutionary plausibility and towards biochemical function. Not only does this seem to be more robust than other preference optimization methods... (1/3)
Functional Alignment of Protein Language Models via Reinforcement Learning with Experimental Feedback 1. This study introduces Reinforcement Learning with Experimental Feedback (RLXF), adapting Proximal Policy Optimization (PPO) and Supervised Fine-Tuning (SFT) to align protein…
Functional alignment of protein language models via reinforcement learning biorxiv.org/content/10.110… #biorxiv_bioeng
Functional alignment of protein language models via reinforcement learning ift.tt/rUYx2oE #biorxiv_bioE
...it works in silico across a wide number of targets, and in vivo! We designed 5 & 10 mutants of CreiLOV - aligned models designed variants with > WT fluorescence 40% of the time, and the best design was a 10 mutant w/ 1.7x fluorescence (maybe the brightest FbFP characterized)
Find out more here⬇️ bioRxiv: biorxiv.org/content/10.110… Code: github.com/RomeroLab/RLXF
A full thread by first author Nathaniel below⬇️
We are excited at the @romerolab1 to share our new preprint introducing RLXF for the functional alignment of protein language models (pLMs) with experimentally derived notions of biomolecular function!