Biology+AI Daily
@BiologyAIDaily
Protein/Antibody Designer, AI for drug design, Deep learning and large language model. Share daily papers on biology + AI
Breaking: The official release of educational guides for AlphaFold Server from @GoogleDeepMind alphafoldserver.com/guides These tutorials aim to assist new users in gaining a deeper understanding of the latest AlphaFold version’s capabilities and maximizing the potential of…

Latent-X: An Atom-level Frontier Model for De Novo Protein Binder Design 1. Latent-X introduces a groundbreaking AI model that generates all-atom protein binders with high affinity and specificity, achieving experimental hit rates exceeding 90% for macrocycles and up to 64% for…
5 months from funding to shipping our first frontier model. Latent-X achieves state-of-the-art hit rates for macrocycles and mini-binders, with picomolar binding affinities — a breakthrough in de novo protein binder design. Available now on our no-code platform!
Latent-X: An Atom-level Frontier Model for De Novo Protein Binder Design 1. Latent-X introduces a groundbreaking AI model that generates all-atom protein binders with high affinity and specificity, achieving experimental hit rates exceeding 90% for macrocycles and up to 64%…
5 months from funding to shipping our first frontier model. Latent-X achieves state-of-the-art hit rates for macrocycles and mini-binders, with picomolar binding affinities — a breakthrough in de novo protein binder design. Available now on our no-code platform!
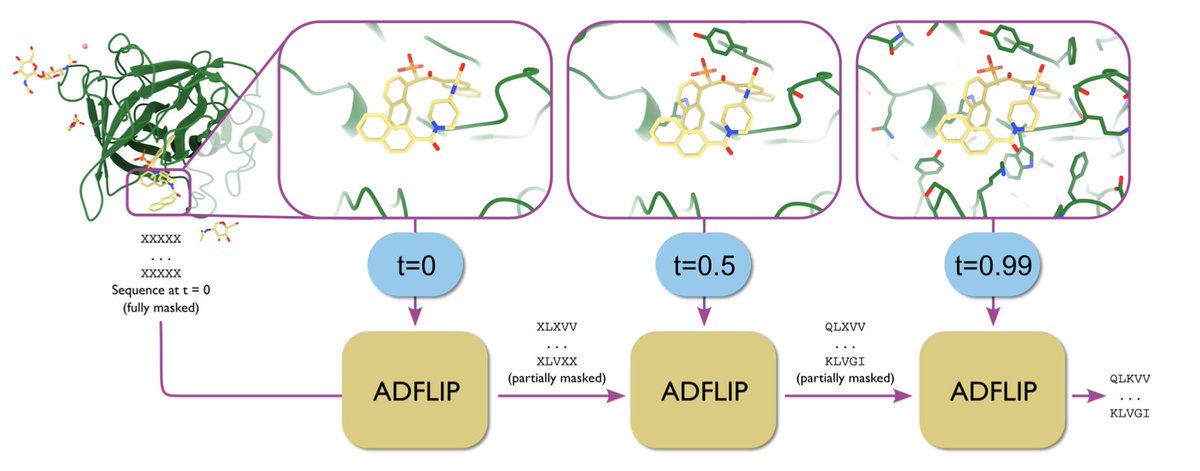
All-atom inverse protein folding through discrete flow matching 1. ADFLIP, a novel generative model for designing protein sequences based on all-atom structural contexts, has been introduced. It addresses the challenges of predicting sequences for complexes with non-protein…
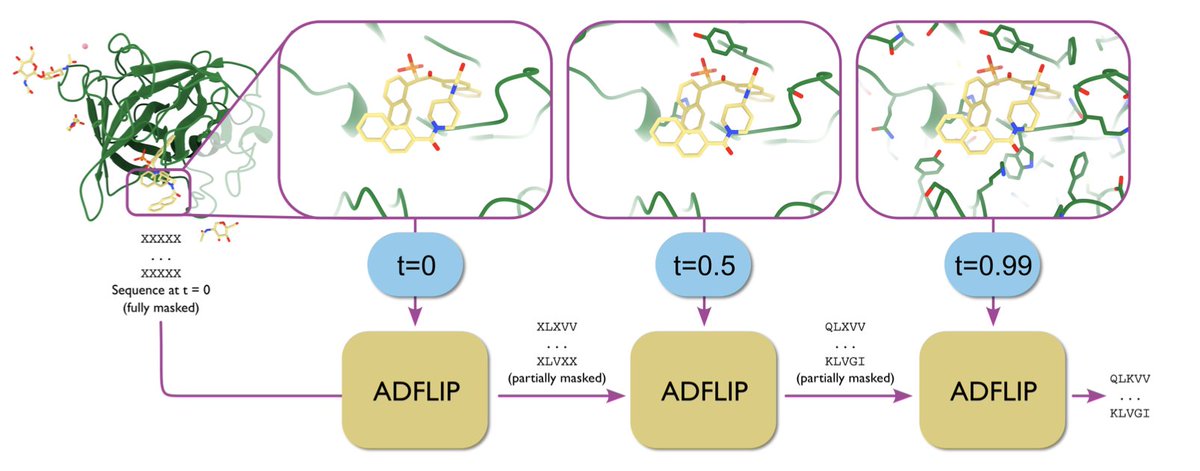
All-atom inverse protein folding through discrete flow matching 1. ADFLIP, a novel generative model for designing protein sequences based on all-atom structural contexts, has been introduced. It addresses the challenges of predicting sequences for complexes with non-protein…
Enhanced Formulation of Precision Probiotics through Active Machine Learning 1. A novel study applies active machine learning (ML) to predict the effects of excipients on the growth of Lactobacillus plantarum, a commonly used probiotic. This approach significantly enhances the…
Enhanced Formulation of Precision Probiotics through Active Machine Learning 1. A novel study applies active machine learning (ML) to predict the effects of excipients on the growth of Lactobacillus plantarum, a commonly used probiotic. This approach significantly enhances the…
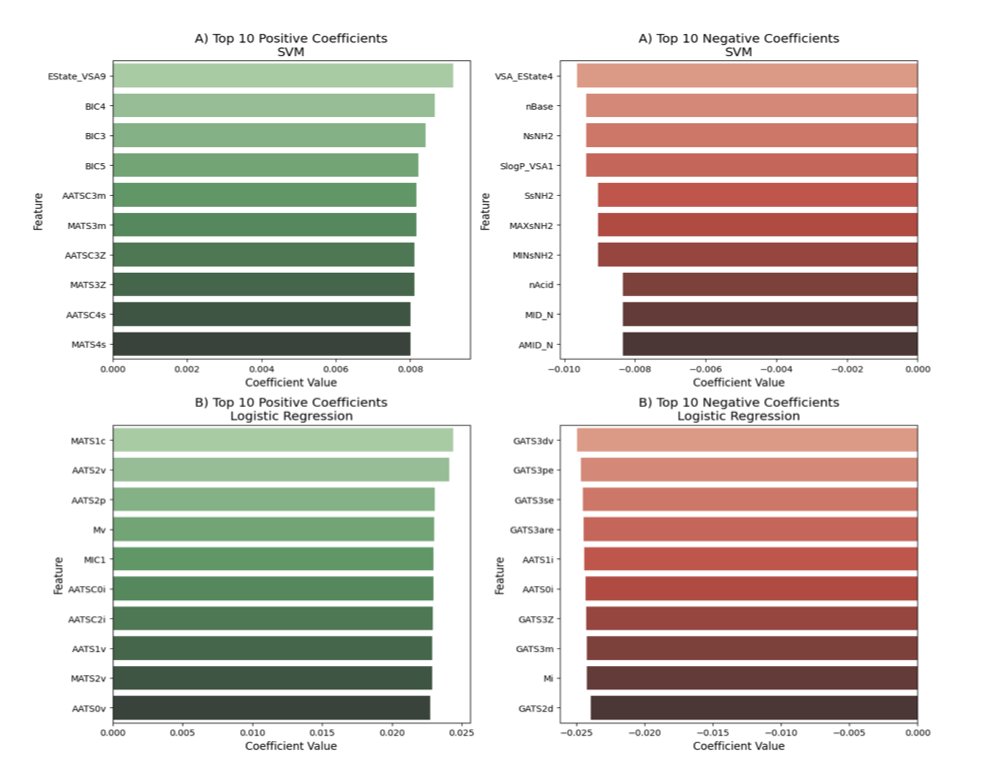
Interventionally-guided representation learning for robust and interpretable AI models in cancer medicine 1. This study introduces a novel class of machine learning models designed for high-dimensional molecular data in cancer medicine, focusing on improving robustness and…

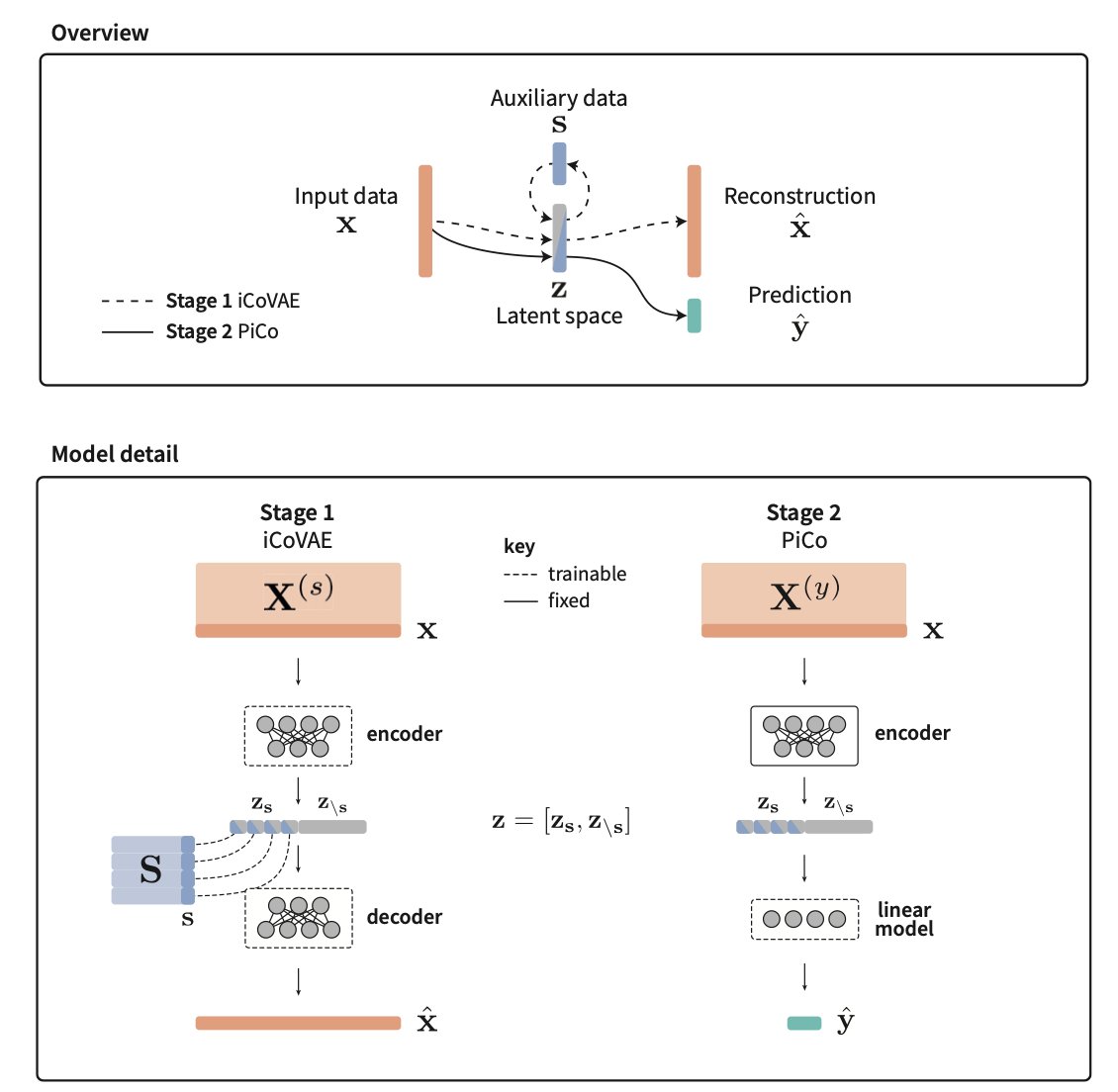
Interventionally-guided representation learning for robust and interpretable AI models in cancer medicine 1. This study introduces a novel class of machine learning models designed for high-dimensional molecular data in cancer medicine, focusing on improving robustness and…
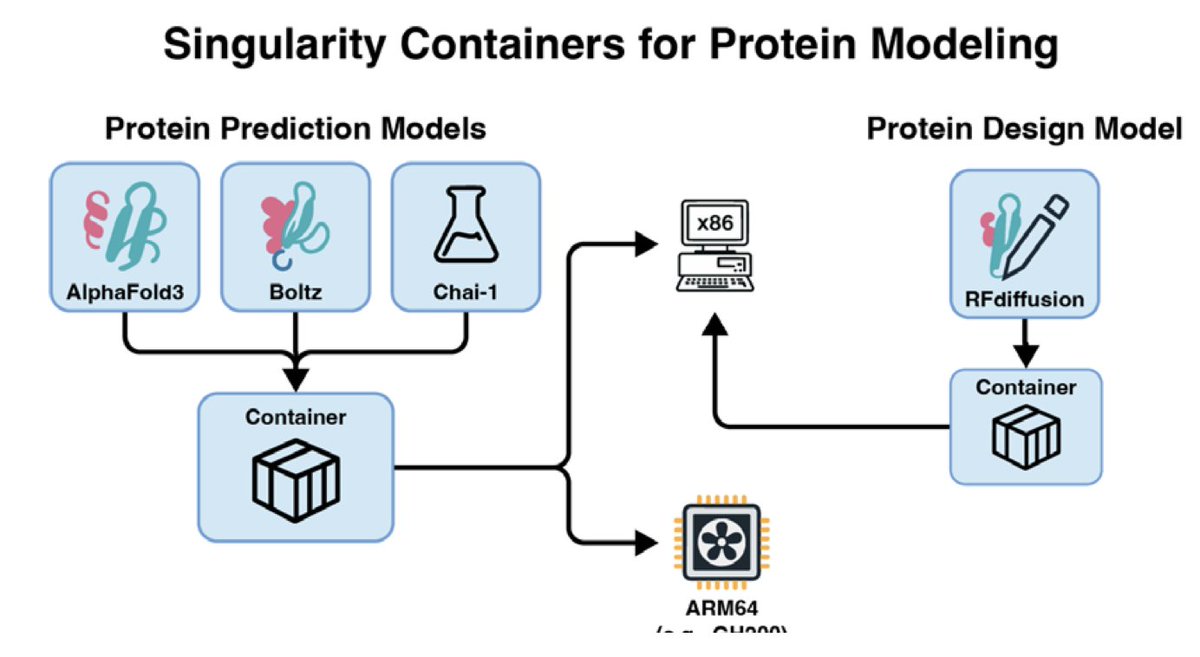
Protein Structure Prediction and Design for High-Throughput Computing 1. Recent advances in structural biology and machine learning have revolutionized molecular biology, enabling accurate protein structure prediction and design at atomic resolution. Tools like AlphaFold3,…
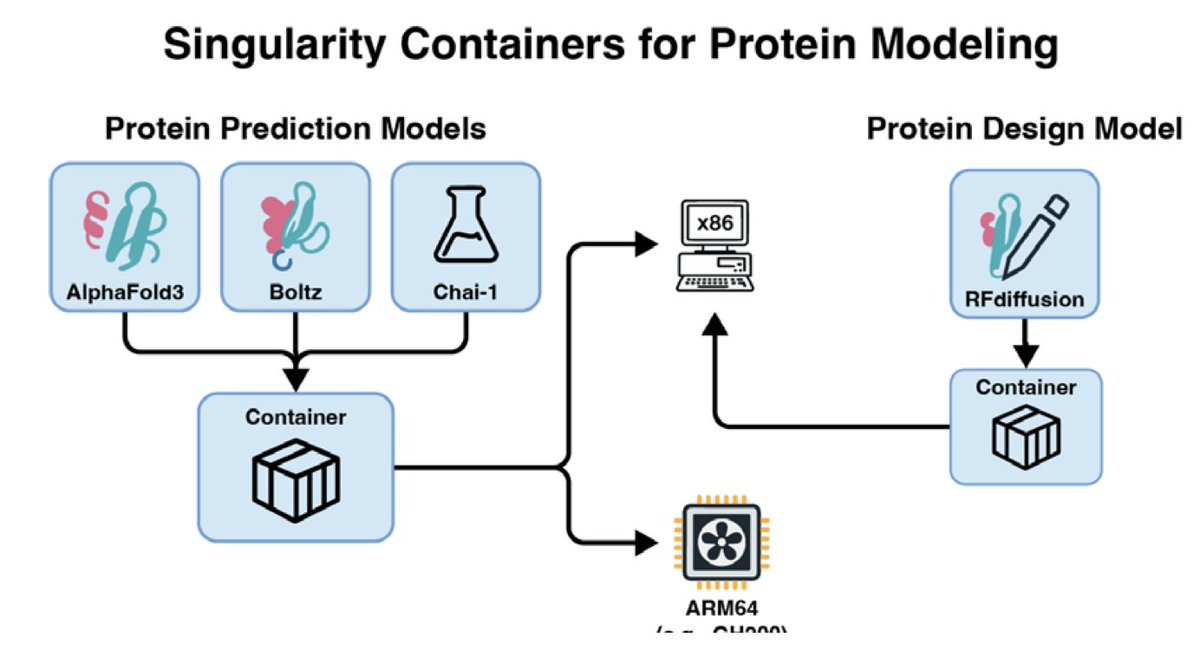
Protein Structure Prediction and Design for High-Throughput Computing 1. Recent advances in structural biology and machine learning have revolutionized molecular biology, enabling accurate protein structure prediction and design at atomic resolution. Tools like AlphaFold3,…
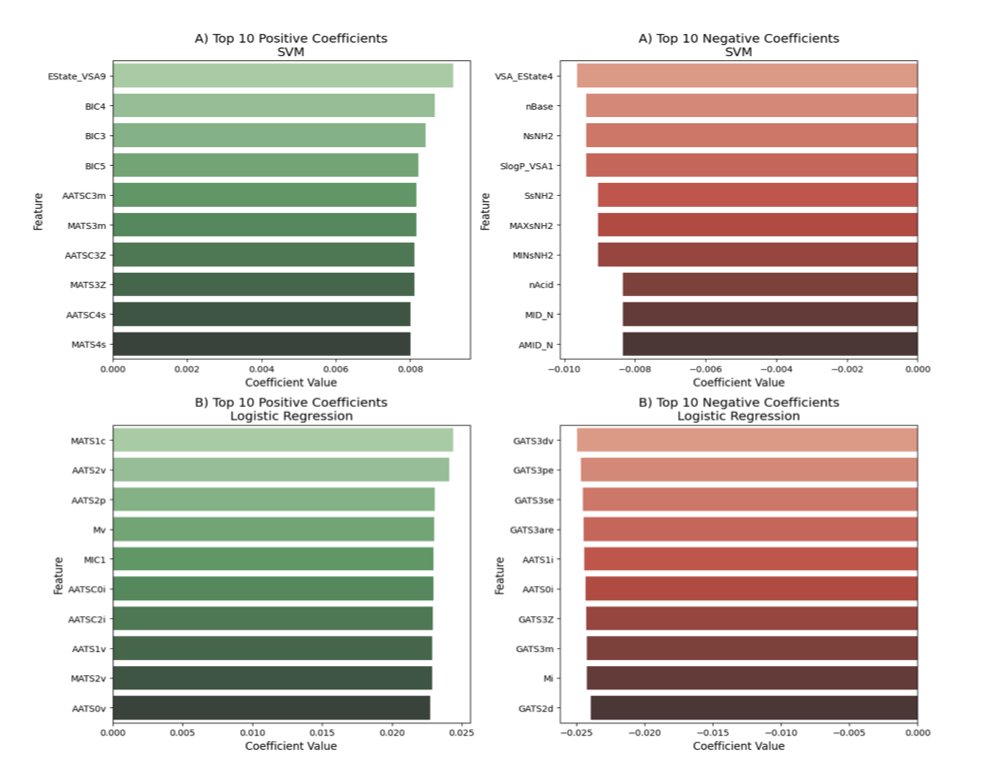
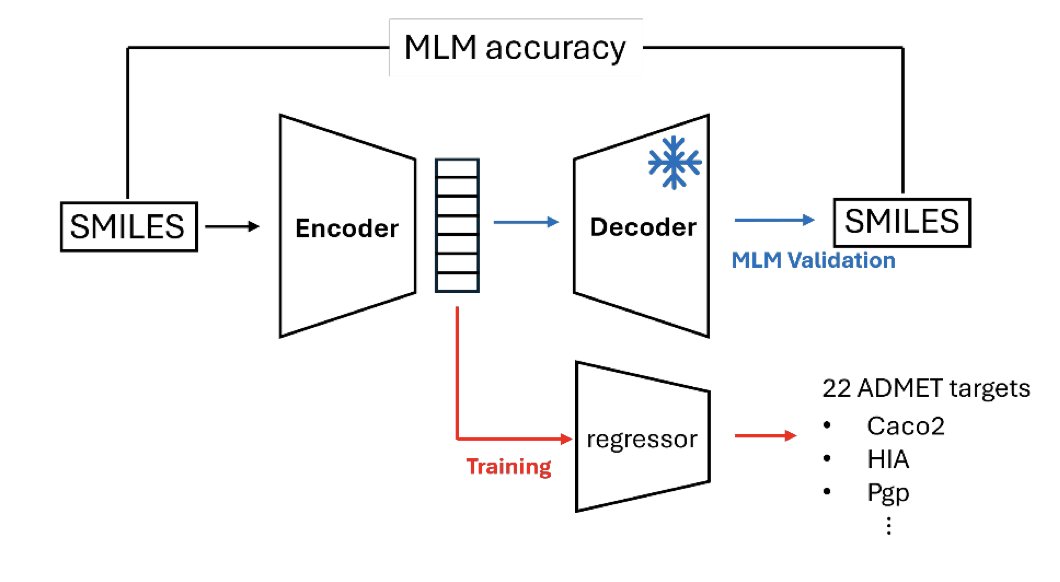
DeBERTa-Based SMILES Encoders for ADMET-Aware Drug Design 1. A new study leverages DeBERTa, a powerful NLP model, to enhance SMILES encoders for predicting ADMET properties in drug design. This approach significantly improves the predictive capacity for critical endpoints like…
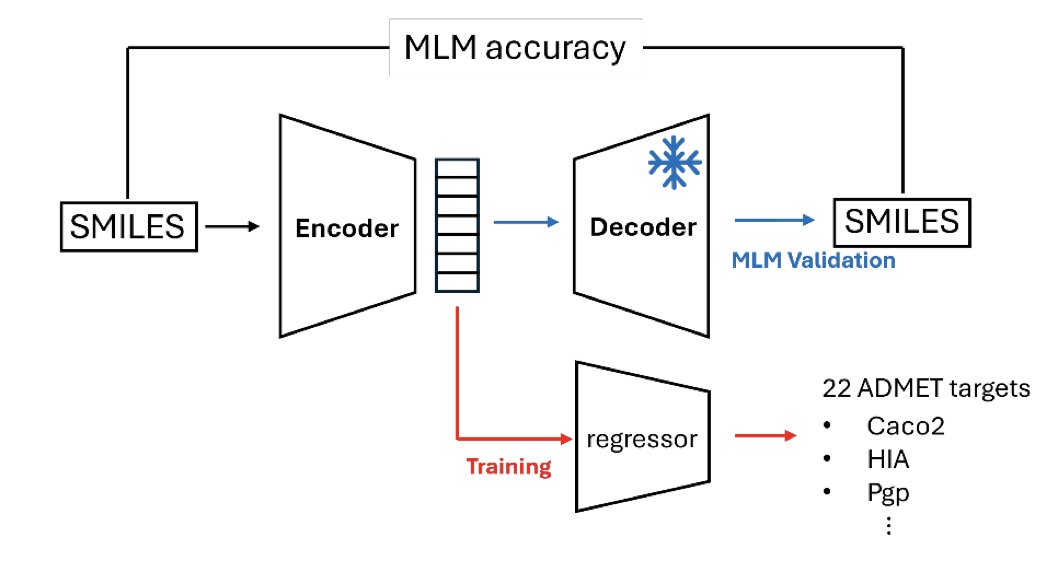
DeBERTa-Based SMILES Encoders for ADMET-Aware Drug Design 1. A new study leverages DeBERTa, a powerful NLP model, to enhance SMILES encoders for predicting ADMET properties in drug design. This approach significantly improves the predictive capacity for critical endpoints like…
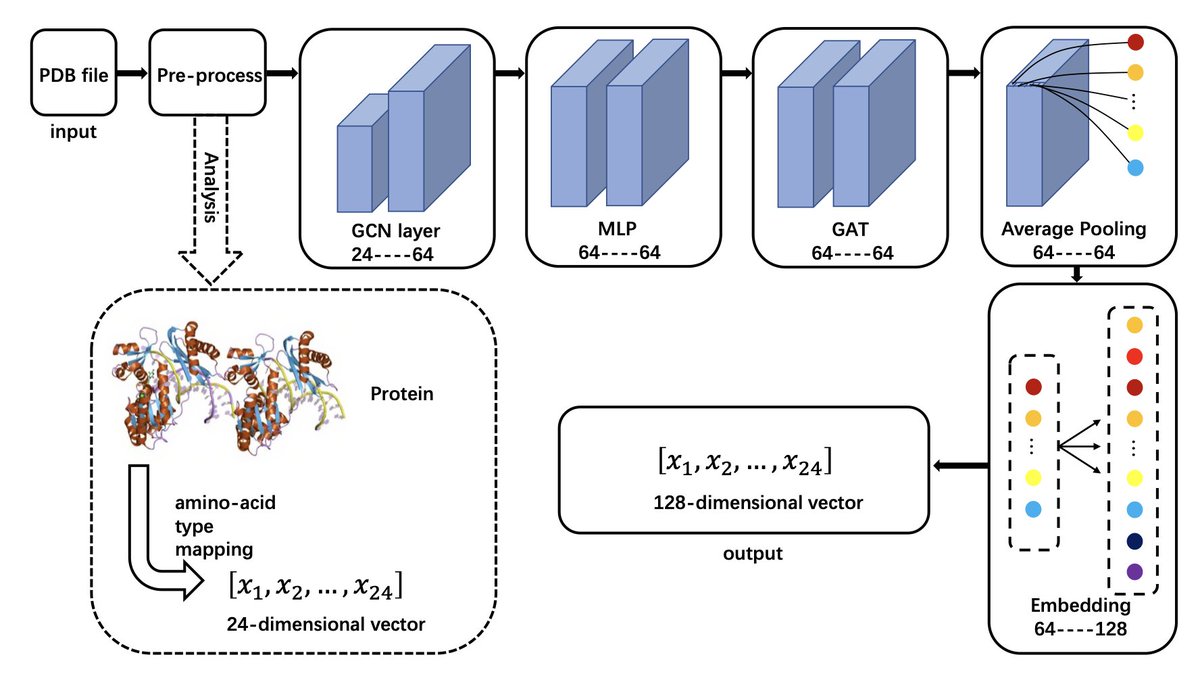
MP-GCAN: a highly accurate classifier for α-helical membrane proteins and β-barrel proteins 1. A novel study in structural bioinformatics introduces MP-GCAN, a novel graph-based classification model designed to accurately distinguish between α-helical membrane proteins, β-barrel…
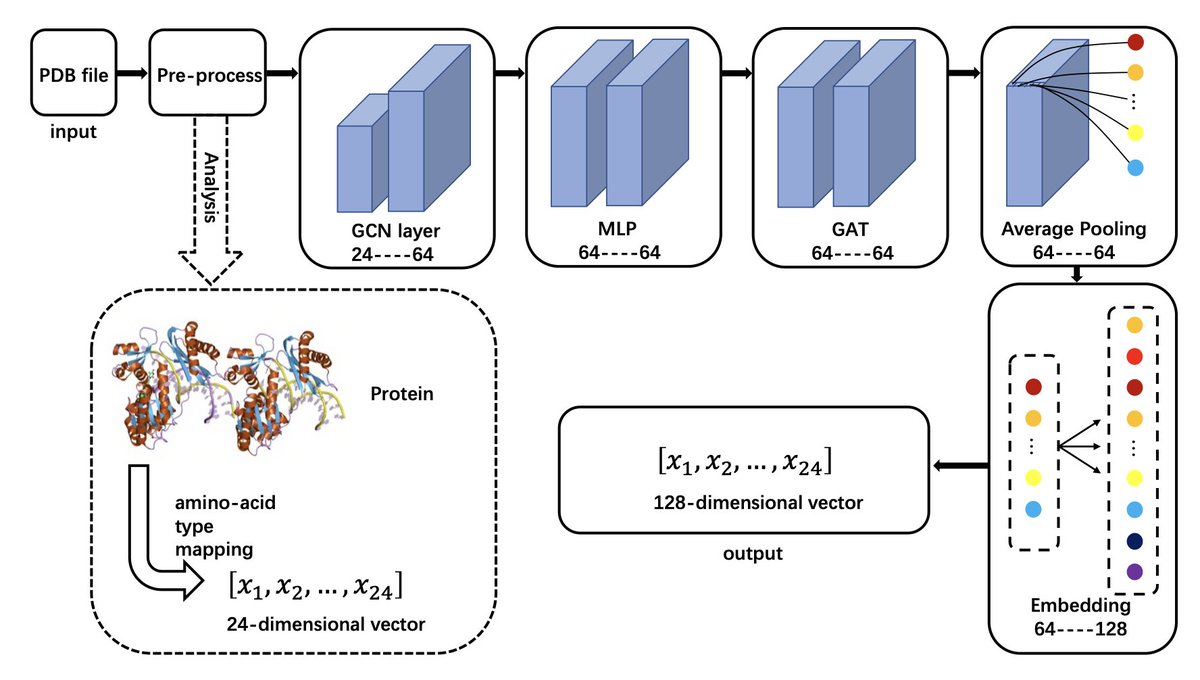
MP-GCAN: a highly accurate classifier for α-helical membrane proteins and β-barrel proteins 1. A novel study in structural bioinformatics introduces MP-GCAN, a novel graph-based classification model designed to accurately distinguish between α-helical membrane proteins, β-barrel…
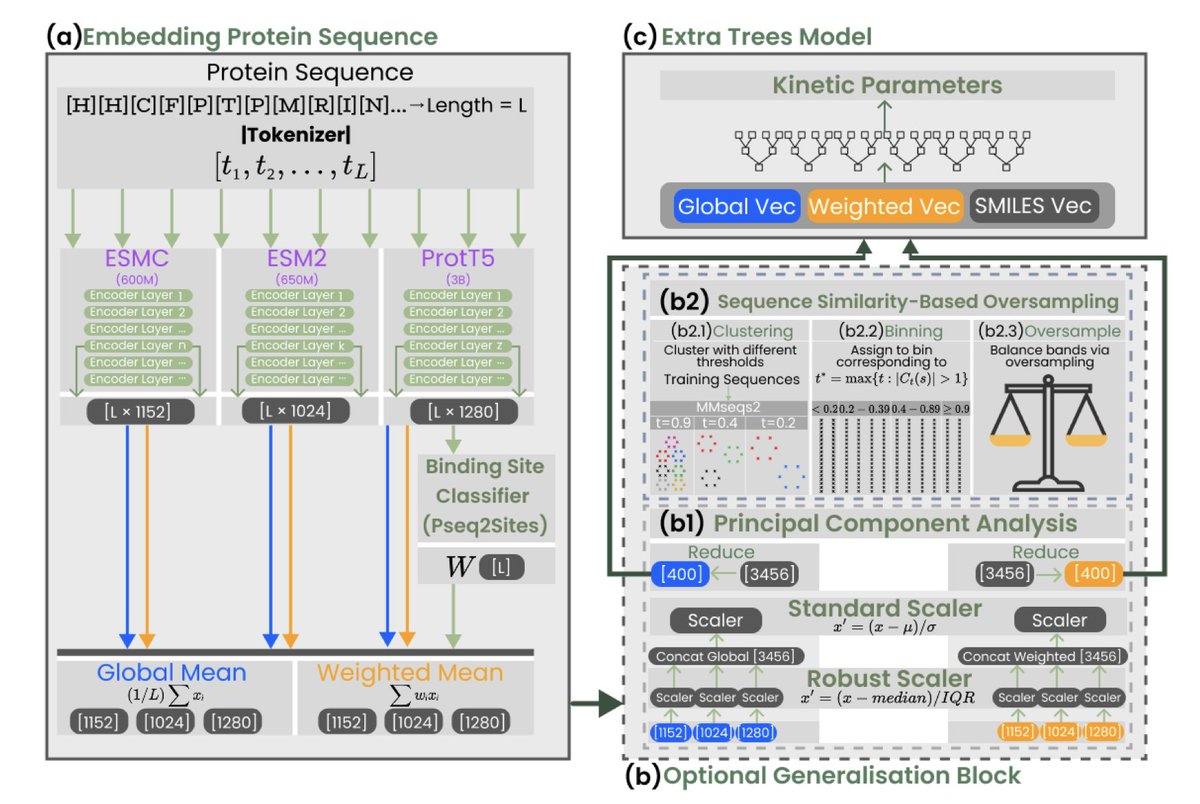
KinForm: Kinetics-Informed Feature Optimised Representation Models for Enzyme kcat and KM Prediction 1. KinForm is a novel machine learning framework designed to improve the prediction of enzyme kinetic parameters kcat and KM. It optimises protein feature representations by…
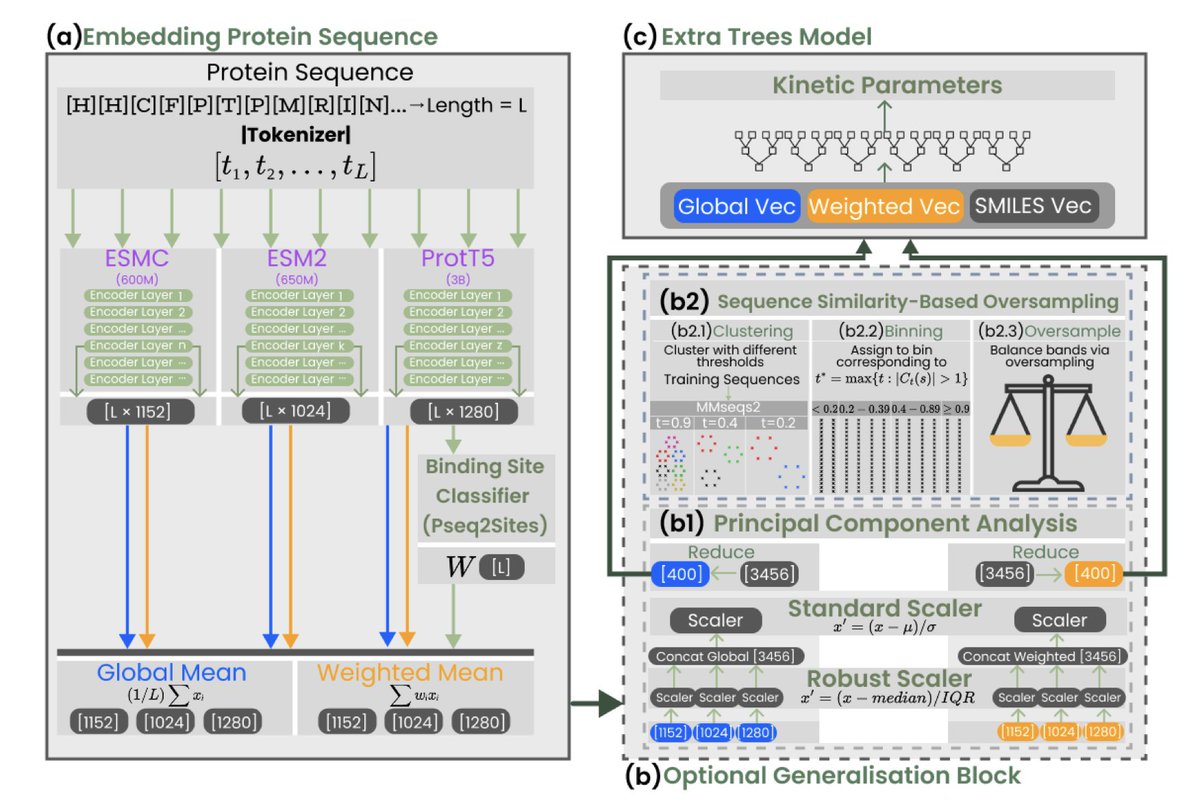
KinForm: Kinetics-Informed Feature Optimised Representation Models for Enzyme kcat and KM Prediction 1. KinForm is a novel machine learning framework designed to improve the prediction of enzyme kinetic parameters kcat and KM. It optimises protein feature representations by…
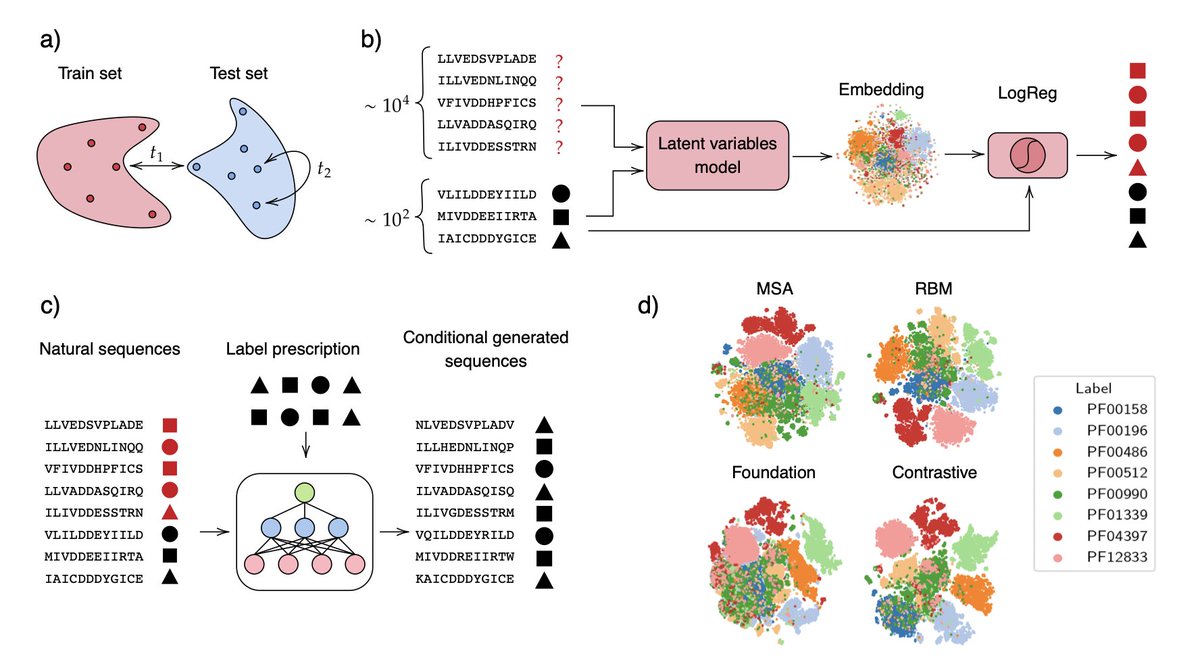
Data augmentation enables label-specific generation of homologous protein sequences 1. This novel study introduces a two-stage approach for semi-supervised functional annotation and conditional sequence generation in protein families using representation learning. The method…
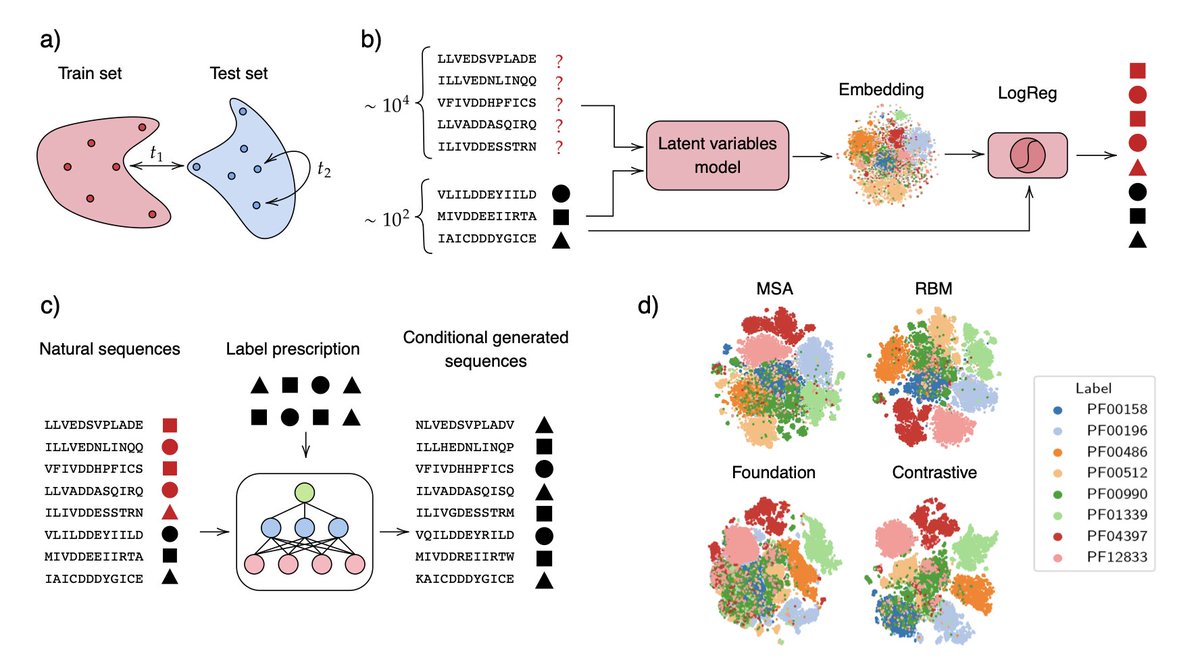
Data augmentation enables label-specific generation of homologous protein sequences 1. This novel study introduces a two-stage approach for semi-supervised functional annotation and conditional sequence generation in protein families using representation learning. The method…