Pierre Boyeau
@pierreboyeau
EECS PhD student @Berkeley working on single cell genomics
Let's pinpoint "The specious art of single-cell genomics" in this tSNE!!!
Our paper "The landscape of biomedical research" is out in @Patterns_CP! Great job by @ritagonmar. cell.com/patterns/fullt… Amazing interactive explorer by @benmschmidt from @nomic_ai: static.nomic.ai/pubmed.html For details see my original Twitter thread: .
What if AI isn’t about building solo geniuses, but designing social systems? Michael Jordan advocates blending ML, economics, and uncertainty management to prioritize social welfare over mere prediction. A must-read rethink. arxiv.org/abs/2507.06268…
Thrilled to be at #ICML this week to talk about #AutoEval I'm particularly interested in model evaluation, single-cell and comp-bio applications! If you're here as well, I'd love to catch up or connect :) arxiv.org/abs/2403.07008
Check out our ICLR workshop on hallucinations, uncertainty, and out-of-distribution performance in foundation models. See you there!
The workshop submission portal is now live: openreview.net/group?id=ICLR.… Looking forward to all the exciting works on hallucinations/uncertainty/out-of-distribution performance on large models. with @SharonYixuanLi @ml_angelopoulos @stats_stephen @barbara_plank @EmtiyazKhan
Announcing ICLR '25 workshop: Quantify Uncertainty and Hallucination in Foundation Models: The Next Frontier in Reliable AI How can we trust large language models (LLMs) when they generate text with confidence, but sometimes hallucinate or fail to recognize their own…
Happy New Year! Our GPN-MSA paper is finally published, under a slightly different title from the preprint. Please check it out and share it with your colleagues. doi.org/10.1038/s41587… 1/4
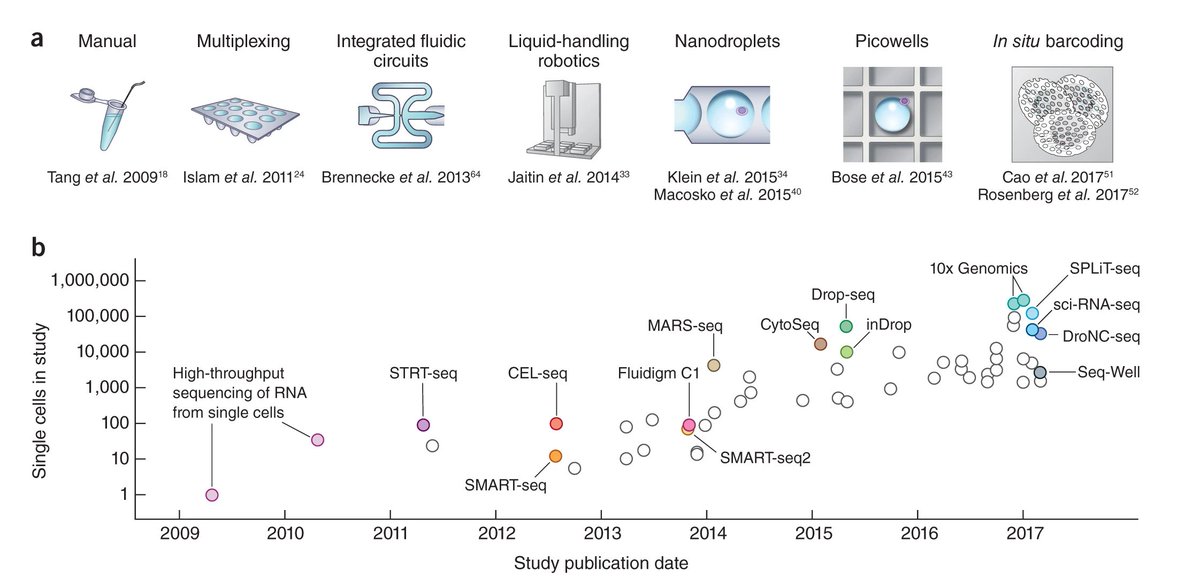
Does anybody know of an updated chart of the evolution of single-cell study/metastudy sizes? @vallens
Trying to install an old version of jax with GPU support with pip:
Had an incredible internship at Altos Labs in Peter Kharchenko's lab in SD ☀ . Thank you for this invaluable and stimulating experience!
We are releasing MrVI: a tool for large-scale multi-sample scRNA analysis. Check out @justjhong tweet for more info!! ⬇️
How can we better reveal cellular 🦠and sample🧍variation from large-scale scRNA-seq studies? We have released a new and improved deep generative model, #MrVI, in scvi-tools and on bioRxiv, accompanied by real-world use cases. A thread... 🧵1/ biorxiv.org/content/10.110…
Happy to announce this work with @AchilleNazaret will be presented at #ICML2024 🥳!
1/🧵I’m excited to share recent work on differentiable causal discovery from @AchilleNazaret and me, supervised by @elhamazizi and @blei_lab. Open the thread for a brief tweetorial (Xtorial?) of our method. arXiv link: arxiv.org/abs/2311.10263
Really enjoyed this collaboration with @rmg2213, Nick Hou, and @joselmcfaline. Please check out if you want to see MrVI in action!
Excited to share a pre-print from the lab describing our effort to use multiplex single-cell chemical transcriptomics to examine differences in tumor cell response associated with the precise manner by which you target an oncogenic kinase. biorxiv.org/content/10.110…
Great talk and shoutout to @FlorianIngelfi1!!
Great talk from @IdoAmitLab on Zman-seq; tackling time in spatial and single-cell genomics #singlecellgenomicsday
Looking forward to it!
Single Cell Genomics Day starts tomorrow at 10AM ET! Come for the awesome keynotes (@JD_Buenrostro , Xiaowei Zhuang, @IdoAmitLab) and to learn about our favorite new methods and trends for the field in the last year. All talks streamed on Youtube and satijalab.org/scgd