Morgan Jones
@mljones1993
Structural biologist working on DNA replication at the @MRC_LMB 🧬
Excited to share my cryo-EM study of human DNA Polymerase Epsilon - mapping interactions with DNA and PCNA, and dissecting how a mismatched base pair is recognised and removed for high fidelity DNA replication! Massive thanks to everyone involved @Yeeles_Lab @MRC_LMB
We are excited to share new mechanistic insights into DNA replication fidelity! How does human DNA Polymerase Epsilon recognise and proofread errors made during synthesis? 🧬 Read @NatureSMB nature.com/articles/s4159…
We are excited to share our latest publication in Nature! Led by our outstanding postdoc, Taha Shahid, & PhD student, Ammar Danazumi, our study reveals how the SV40 LTag helicase melts and translocates on DNA, driven by an entropy-switch mechanism. nature.com/articles/s4158…
Great new work from the very talented Emma Fletcher (@Yeeles_Lab). Biochemistry, and a touch of cryoEM, reveal why the DNA replication machinery needs a dedicated leading strand PCNA clamp loader. 🧬🧪🥼🔬🧑🔬 embopress.org/doi/full/10.10…
The first paper from our lab is online! A wonderful collaboration with @scottbscience lab. We discover that Pol II is extensively regulated by Ubiquitin at the beginning of the transcription cycle! 🧵 cell.com/molecular-cell…
NEW from us @TheCrick! Capture, mutual inhibition and release mechanism for aPKC-Par6 and its multi-site polarity substrate Lgl | bioRxiv #Science #Research #StructuralBiology #CryoEM #Crystallography #Polarity #Cancer biorxiv.org/content/10.110…
Congratulations to @MaikEngeholm , @LucasFarnung and everyone involved at @mpgpresident Lab. Maik was the first person to introduce me in detail to cryo-electron microscopy and image processing in RELION and it was a great experience to learn from him!
Online Now: Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT dlvr.it/TD8qRV
🚨📣 Excited to share our latest work investigating how the replisome responds to topoisomerase-DNA crosslinks 🧬🔬 cell.com/molecular-cell… Fantastic work from @rosewesthorpe and @JohannJRoske 🍄🍄🍄🍄
Amazingly—the day after I posted my video exploring their structures of the human replisome—the @Yeeles_Lab and @JohannJRoske put out a BEAUTIFUL paper featuring the autocorrect function inside DNA Polymerase Epsilon. Quick loop of that data: paper here: go.nature.com/4dDjjZd
Brilliant work from super talented PhD student @JohannJRoske. Amazing how he managed to identify such tiny but key conformational changes that are critical to the mechanism
We are excited to share new mechanistic insights into DNA replication fidelity! How does human DNA Polymerase Epsilon recognise and proofread errors made during synthesis? 🧬 Read @NatureSMB nature.com/articles/s4159…
We are excited to share new mechanistic insights into DNA replication fidelity! How does human DNA Polymerase Epsilon recognise and proofread errors made during synthesis? 🧬 Read @NatureSMB nature.com/articles/s4159…
This is so cool! @this_clockwork made animations of eukaryotic DNA replication using our structures! Watching similar vids of dynein on microtubules sparked my interest in structural biology, and now our work is used in a similar way. Great job! #StructuralBiology #DNAReplication
Here's a render of a full eukaryotic replisome carrying out DNA replication. This was built using real .pdb data from @buildmodels I used this to make my latest video: (youtu.be/lv89fSt5jBY) But now I want to do a quick works cited 🧵 (1/ ?)
Here's a render of a full eukaryotic replisome carrying out DNA replication. This was built using real .pdb data from @buildmodels I used this to make my latest video: (youtu.be/lv89fSt5jBY) But now I want to do a quick works cited 🧵 (1/ ?)
Our paper looking at the structure of retrograde IFT trains is out now! Have a look at the link, or see some of the headlines in this video: cell.com/cell/fulltext/…
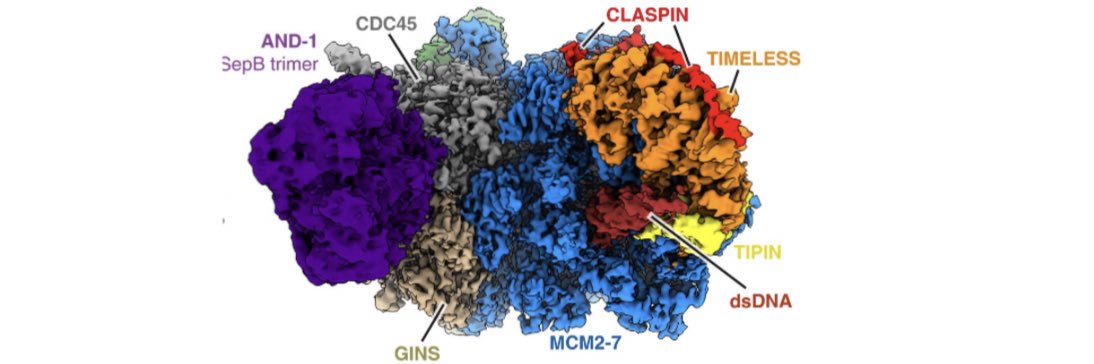
Excellent work from the Deegan lab @TDDeego @mrc_hgu! They very nicely show how different accessory helicases are recruited to the replisome, with key implications for replication termination and the bypass of obstacles on the DNA. Congratulations!
New paper from my group, in which we use the combined power of AlphaFold and biochemical reconstitution to determine how the Rrm3 helicase is targetted to the eukaryotic replisome embopress.org/doi/epdf/10.10…
New paper from my group, in which we use the combined power of AlphaFold and biochemical reconstitution to determine how the Rrm3 helicase is targetted to the eukaryotic replisome embopress.org/doi/epdf/10.10…
Brilliant to see our talented postdoc, Cristian, publish his work on CMG disassembly in @embojournal. The culmination of his PhD work in the Labib lab at @dundeeuni. @TDDeego embopress.org/doi/full/10.10…..
Put together a model of the full human replisome/ primosome complex using the GORGEOUS CryoEM data deposited at @buildmodels by @Yeeles_Lab and researchers like @mljones1993 Completely blown away by the level of detail structural biologists are able to render these days
CMG helicase disassembly is essential and driven by two pathways in budding yeast biorxiv.org/cgi/content/sh… #bioRxiv
Please share & repost! Thanks! Are you excited about neuroimmunology? Do you have/are you finishing a PhD in neuroscience, (neuro)immunology or organoid biology? I'm recruiting a postdoc to join me this September/October at the new lab! @MRC_LMB nature.com/naturecareers/…
Parental histone transfer caught at the replication fork nature.com/articles/s4158…