Maya Arce
@maya_m_arce
Excited to share the latest from the @MarsonLab! An investigation of context-specific gene regulation in T cells, which led to dynamic mechanisms required to control T cell rest and activation. Published today in @Nature thanks to the work of many! nature.com/articles/s4158…
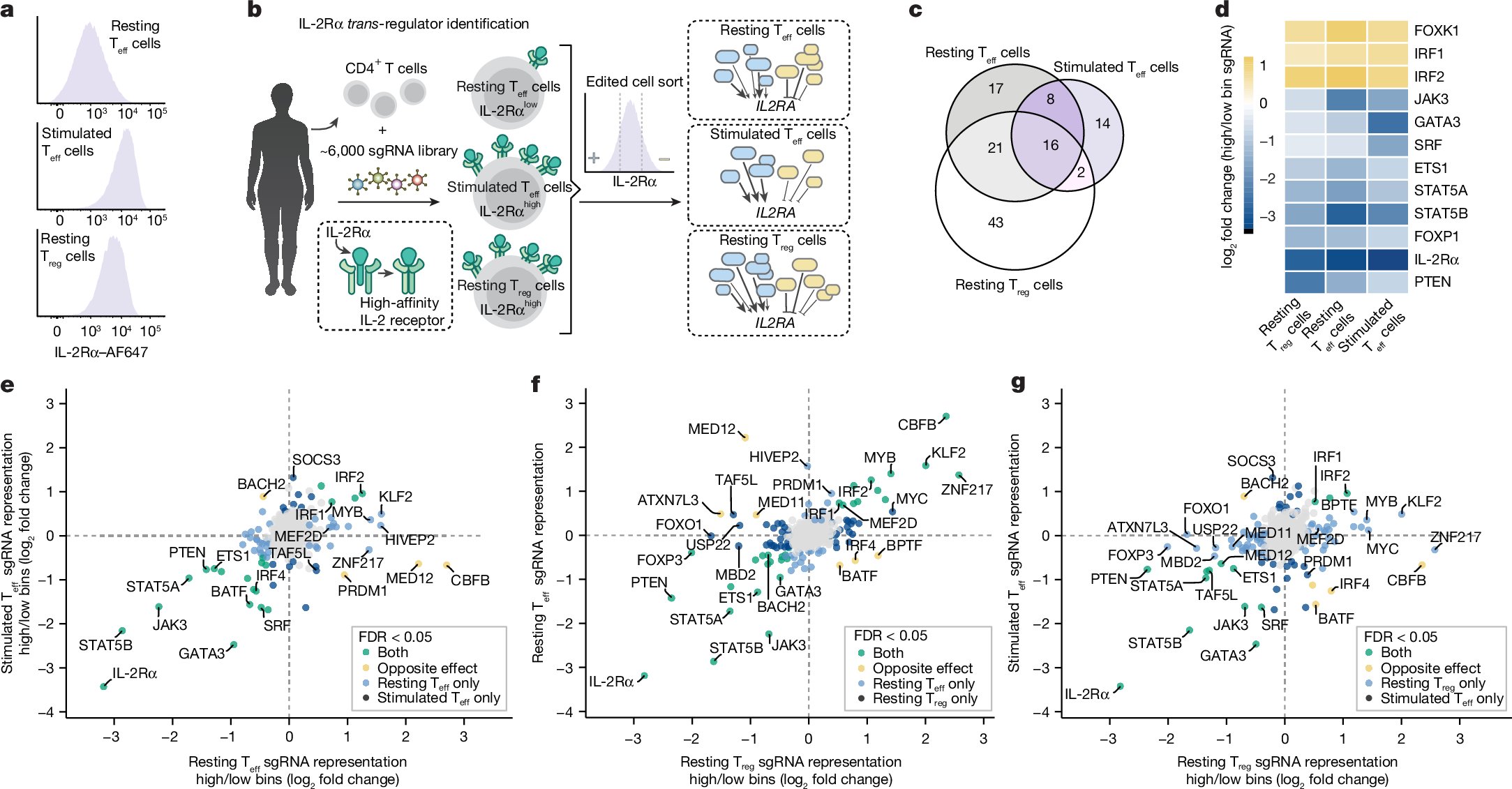
Thank you Jonathan for the elegant explanation of our recent work! We dove into how we can model the gene regulatory architecture of complex traits with 1) gene effects from LoF burden tests and 2) Perturb-seq. It’s been an incredibly exciting and thought-provoking journey!
Modern GWAS can identify 1000s of hits but it can be hard to turn this into biological insight. What key cellular functions link genetic variation to disease? I'm excited to present our new work combining associations + Perturb-seq to build interpretable causal graphs! A🧵
Think of a conductor guiding an orchestra—scientists discovered how T cells fine-tune their responses to keep the immune system in harmony. @MarsonLab @maya_arce_ @KroganLab @Satpathology @jkpritch @Nature @UCSF @GladstoneInst nature.com/articles/s4158…
A team of scientists uncovered how one protein controls the behavior of immune cells, which could have applications for treating cancer and autoimmune conditions. @UCSF @QBI_UCSF @parkerici @MarsonLab @maya_arce_ @KroganLab @Satpathology @jkpritch @Nature bit.ly/4f9lGTP
Thrilled to share our work on inferring gene networks in T cells using CRISPR perturbations. We show how to build these networks + use them to interpret GWAS. This work co-led with @maya_arce_ + senior authors @jkpritch @alexisjbattle @MarsonLab 🧵👇tinyurl.com/2spd6hwu