
Jason Zhaoxing Zhang
@jasonzxzhang
Molecular tool designer for signaling | Assistant Professor at @UCLA | 🏳️🌈🇺🇸 not on Twitter, on BlueSky: https://bsky.app/profile/jasonzxzhang.bsky.social
I am transitioning my social media presence from here to Bluesky. For me, Twitter is no longer a reliable source of information and I vehemently despise Elon. If you want to be up to date with what I do, then feel free to add me on Bluesky: bsky.app/profile/jasonz…
Online now! De novo designed Hsp70 activator dissolves intracellular condensates. Check out the study by chemistry laureate @Uwproteindesign David Baker, @jasonzxzhang & others in @CellChemBiol #bps2025 hubs.li/Q036KN030
Excited to share my recent work on designing new Hsp70 activators to regulate condensates, and thus reveal their functions: sciencedirect.com/science/articl… @UWproteindesign
Just started my BlueSky account: jasonzxzhang.bsky.social Feel free to add me there as well! Will see how this goes 🤔
This is an honor well deserved! I am always amazed by the sheer ingenuity David and our lab always produces. Congratulations to David! @UWproteindesign
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”
Fantastic first day of science with our Postdoc Rising Stars and amazing keynote by @TheVillaLab on jumbo phages!@UofUBiochem @uofunuip @huntsmancancer @uofupharmacy @BWFUND
Excited to visit @UUtah! I really appreciate the opportunity to share my recent work @sigalalab @Myers_lab 🙏🏼!
We're excited to announce the upcoming Postdoctoral Rising Stars Symposium @UofUHealth with 20 outstanding scientists from broad backgrounds and keynote by @TheVillaLab, with generous support by @BWFUND. Hosts @UofUBiochem @uofunuip @UUNeurobiology @uofupharmacy @huntsmancancer
Thrilled to share progress on Ras isoform binders! Using AI-driven protein design, I’ve targeted the challenging, highly charged and disordered C-terminus of Ras. These de novo binders exceed antibody specificity and effectively disrupt Ras signaling. biorxiv.org/content/10.110…

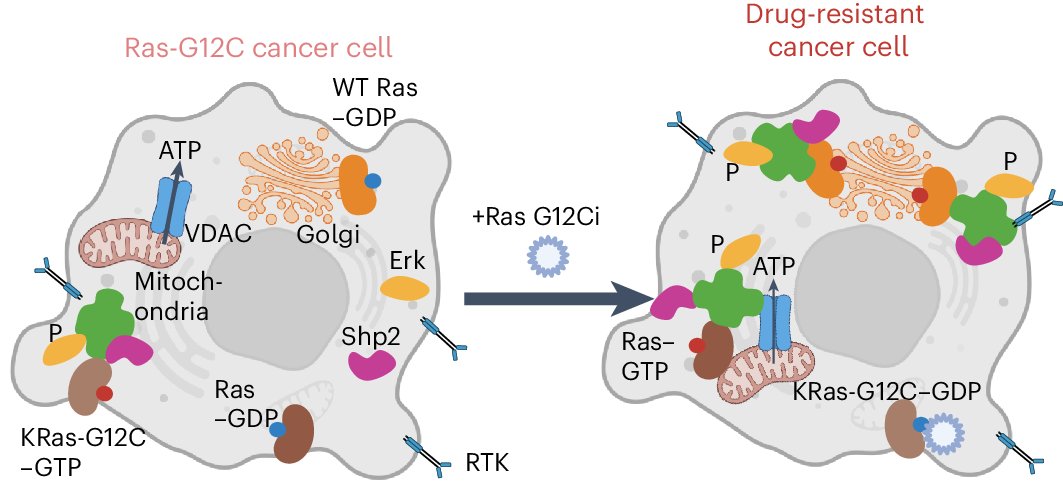
Recently out at Nat Chem Bio! I used my Ras biosensors (Zhang et al Nat Biotech 2024) to profile the activity and environment of Ras at the single-cell level, which unveiled the signaling mechanisms underlying RasG12C inhibitor resistance. nature.com/articles/s4158… @UWproteindesign
Reason to go to conferences: collect their cute shirts 😂 @ProteinSociety #PS38

Excited to share that my K99 at NCI is awarded! I will continue to build new protein tools to understand the signaling programs enabling Ras drug resistance. And with that, a shameless plug that I’m on the faculty market this Fall 2024 👀.

The future faculty page of Baker lab postdocs is now online! bakerlab.org/future-faculty/ Please check out the cool things my colleagues and I are doing and plan to do!
Announcing AlphaFold 3: our state-of-the-art AI model for predicting the structure and interactions of all life’s molecules. 🧬 Here’s how we built it with @IsomorphicLabs and what it means for biology. 🧵 dpmd.ai/3URDiNo
NCI blog post about my de novo designed Ras biosensors: cancer.gov/research/key-i…

Giving a talk at CSHL on my 30th birthday 😬
Happy to share that the Ras sensors (Ras-LOCKR-S and Ras-LOCKR-PL) are now on Addgene! addgene.org/browse/article…
All atom biomolecular modeling 🤯 @UWproteindesign science.org/doi/10.1126/sc…
Protein structure design is here! Hope our recent work (nature.com/articles/s4158…) on de novo design of protein sensors will be interesting and relevant to others curious in functional protein design.
Spotlight on protein structure design go.nature.com/48jEDjr
